Reviews:
Cell Stress, Vol. 2, No. 5, pp. 96 - 112; doi: 10.15698/cst2018.05.134
Putting together and taking apart: assembly and disassembly of the Rad51 nucleoprotein filament in DNA repair and genome stability
1 Institute of Cell Biology, School of Biological Sciences, University of Edinburgh.
Keywords: homologous recombination, Rad51 filament, double-stranded DNA break, DNA repair, Rad51 regulation.
Abbreviations:
BIR – break-induced replication,
DNTA – de novo telomere addition,
DSB – double-stranded DNA break,
dsDNA – double-stranded DNA,
GC – gene conversion,
HR – homologous recombination,
PTM – post-translational modification,
RFC – Replication Factor C,
RPA –Replication Protein A,
SDSA – synthesis-dependent strand annealing,
SIM – SUMO-interacting motif,
SSA – single-stranded annealing,
ssDNA – single-stranded DNA.
Received originally: 01/02/2018 Received in revised form: 12/03/2018
Accepted: 13/03/2018
Published: 28/03/2018
Correspondence:
Svetlana Makovets, Institute of Cell Biology, School of Biological Sciences, University of Edinburgh, Alexander Crum Brown Road, King’s Buildings, Edinburgh, EH9 3FF, United Kingdom Sveta.Makovets@ed.ac.uk
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Tadas Andriuskevicius, Oleksii Kotenko and Svetlana Makovets (2018). Putting together and taking apart: assembly and disassembly of the Rad51 nucleoprotein filament in DNA repair and genome stability. Cell Stress 2(5): 96-112. doi: 10.15698/cst2018.05.134
Abstract
Homologous recombination is a key mechanism providing both genome stability and genetic diversity in all living organisms. Recombinases play a central role in this pathway: multiple protein subunits of Rad51 or its orthologues bind single-stranded DNA to form a nucleoprotein filament which is essential for initiating recombination events. Multiple factors are involved in the regulation of this step, both positively and negatively. In this review, we discuss Rad51 nucleoprotein assembly and disassembly, how it is regulated and what functional significance it has in genome maintenance.
INTRODUCTION
Homologous recombination (HR) involves exchange of genetic information, often between two different DNA molecules. This exchange requires physical interaction between the molecules and may lead to heritable genetic changes, contributing to biodiversity and higher evolutionary adaptability of species. Novel combinations of alleles arising in meiosis is a great example of genetic variability as a result of HR. On the other hand, physical linkage between chromosomes during meiotic HR is important for pairing and accurate segregation of homologous chromosomes in meiosis, thereby preventing aneuploidy and promoting genome integrity. In addition, HR machinery contributes to genome stability by playing a central role in stabilisation and restart of stalled replication forks as well as repair of DNA breaks routinely arising from broken replication forks, oxidative damage, etc. Because DNA breaks are part of normal cell physiology, unsurprisingly, defects in HR in humans lead to developmental disorders and cancer predisposition [1].
–
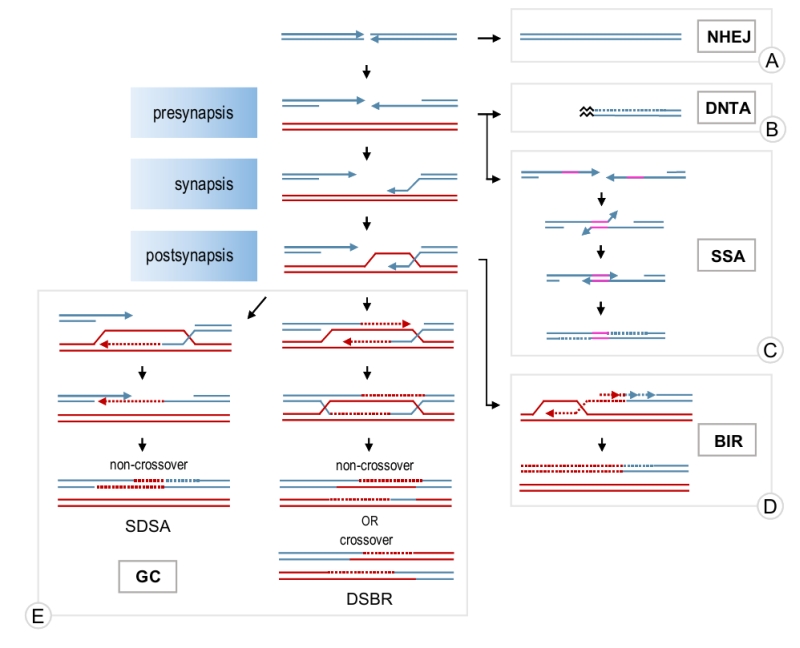
A double-stranded DNA break (DSB) is often considered the most dangerous type of DNA damage as it disrupts the continuity of chromosomes and, if unrepaired, may lead to loss of genetic information and eventual cell death. Two major mechanisms are normally used to repair a DSB: non-homologous end joining (NHEJ) and HR (Figure 1). NHEJ involves ligation of the broken ends with little or no DNA processing around the break. It is efficient but often mutagenic due to small deletions or insertions at the damage locus [2][3][4]. Occasionally, it can also lead to gross chromo-somal rearrangements due to illegitimate joining of DNA ends from different breaks or ligations of chromosome breaks to telomeres [5][6][7]. HR includes a group of pathways sharing two features: i) they require intact homologous DNA sequences for the repair (called donor DNA) and ii) they stem from the same original step of extensive DSB resection around the break, which generates single-stranded DNA (ssDNA) necessary for HR pathways to operate. Although DSB repair is well-conserved among eukaryotes, the preference for the repair mechanism is cell cycle-dependent and species-specific [3][8][9]. Occasionally, a DSB can be mistakenly recognised as a telomere and telomerase, the enzyme responsible for telomere extension, heals the break by adding a de novo telomere (Figure 1) [10]. Although de novo telomere addition (DNTA) may stabilise the broken chromosome, it results in terminal deletions.
–
–
Depending on the nature of a DSB break, homology-dependent repair may proceed through gene conversion (GC), break-induced replication (BIR) or single-strand annealing (SSA; Figure 1). GC is used to repair a DSB when both ends of the break have homology to a donor DNA molecule and are available for the repair, whereas BIR is employed when only one end is present, for example, to restore broken replication forks [11][12]. SSA does not involve external homology and can only be used if a DNA break is flanked by homologous repeats in direct orientation. SSA is mutagenic as any sequences between the homologies and one of the two copies of the repeated sequences are lost [13]. However, SSA may act as a safeguard mechanism when other options are not available.
–
All three homology-dependent repair pathways begin with the resection of the 5’ end around a DSB to create ssDNA overhangs [14]. SSA may occur at this point through annealing of the complementary sequences on the resected ends [13]. GC and BIR require invasion of the resected DNA end into a donor molecule and a subsequent strand exchange where one strand of the donor molecule is displaced by the invading strand to form a D-loop. The invading strand is then extended using the homologous donor as a template [11][12]. In the case of GC, only the area around the DSB is copied. GC may then proceed through a so-called synthesis-dependent strand annealing (SDSA) mechanism where the invading strand is displaced from the donor molecule and annealed to the sequences on the other end of the break (Figure 1). Alternatively, the second end of the break may be captured by the D-loop forming a double Holliday junction intermediate which can then be resolved by endonucleases or helicases (double-strand break repair pathway – DSBR) [11]. In contrast to SDSA, which generates exclusively non-crossovers, the resolution of a double Holiday junction may lead to either a crossover or a non-crossover. In BIR, the donor molecule is replicated from the invasion site all the way to the telomere by conservative replication (Figure 1) [15]. Both, GC and BIR may result in error-free repair if a sister chromatid is used as a donor. However, if the repair involves a homologous chromosome or an ectopic homology site, loss of heterozygosity or chromosomal rearrangements may occur [11][12][16].
–
The homology search and strand exchange reaction required for DSB repair by GC and BIR are catalysed by a recombinase called Rad51. Rad51 binds to resected ssDNA forming a Rad51-ssDNA filament (presynapsis) which then catalyses the search for homologous sequences and establishes a physical contact between the broken and donor molecules (synapsis) by invading the duplex donor DNA (Figure 1). As this step is absolutely required and is often rate-limiting in DSB repair, Rad51 assembly and disassembly plays an important role in modulating HR. In addition, disassembly of Rad51 filament is also required at the late stages of repair to restore the double-stranded structure of DNA [17]. Recently, a novel role for Rad51 in protection and restart of stalled replication forks has emerged [18]. Therefore, understanding the dynamics of Rad51 filaments is important for elucidating the molecular mechanisms of DNA repair and genome stability.
RAD51 RECOMBINASE
Rad51 was first genetically identified in 1974 in a screen for Saccharomyces cerevisiae mutants sensitive to ionising radiation [19]. Since then, the involvement of Rad51 in genome integrity and dynamics has been well-characterised, defining it as one of the key enzymes required for HR [20][21][22][23][24][25]. Rad51 is conserved among eukaryotes: Rad51 from vertebrates shares on average 74% protein sequence identity with fungi and plants while the human and mouse homologs are 99% identical [26]. Rad51 belongs to the ancient RecA/RAD51 protein family which apart from the bacterial and archaeal orthologues, RecA and RadA respectively, includes members that have diverged from the original function and adapted to more specialised roles. For example, Dmc1 is a meiosis-specific equivalent of Rad51, whereas yeast Rad55 and Rad57 are Rad51 paralogues involved in Rad51 regulation [27][28][29][30]. Although most eukaryotic orthologues are called Rad51 some, like the Schizosaccharomyces pombe orthologue Rhp51, may have a different name. For simplicity, these orthologues as a whole will be referred to as Rad51/RAD51 further in the review.
–
High protein sequence conservation suggests that the function of Rad51 is also conserved. Indeed, Rad51 deficient yeast and mouse trophoblast-like cells exhibit closely similar defects, including increased sensitivity to ionising radiation and chromosome loss, both of which can be explained by compromised DSB repair [19][25][31]. However, mammalian cells appear to be more sensitive to the loss of RAD51: a homozygous gene deletion results in embryonic lethality and failure to establish Rad51 null cell lines [31]. This is also true for the chicken DT40 cells which accumulate spontaneous DNA DSBs in the absence of RAD51 and eventually die [32]. The stronger severity of the RAD51 null phenotype in vertebrates compared to yeasts suggests a greater reliance of these cells on conserved RAD51 activities, possibly due to the emerging role of RAD51 in preventing DNA damage during conventional replication [18] or the higher frequency of stochastic DSBs per cell cycle stemming from the larger genome sizes [31].
–
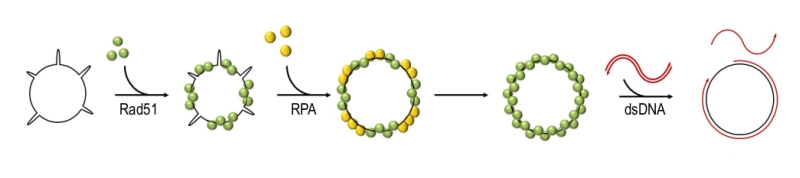
Rad51 exists as a monomer in a solution but it can polymerise on both double-stranded DNA (dsDNA) and ssDNA in a cooperative manner and forms a right-handed nucleoprotein filament, in which the DNA is stretched and extended to facilitate homology search and base pairing [24][33][34][35][36][37]. Unlike RecA, which has a very low affinity for dsDNA, Rad51 can bind both ssDNA and dsDNA in vitro, albeit with a preference for ssDNA [24][38][39][40]. As required for its recombinase activity, Rad51 can bind two DNA molecules simultaneously, through its primary and secondary DNA binding sites. The primary binding site is responsible for the interactions with DNA during the formation of the filament and the double-stranded product in postsynapsis, while the secondary site is required for the capture of a donor molecule in presynapsis [41][42][43]. Rad51 can carry out the strand-exchange reaction in vitro (Figure 2). This activity is strongly stimulated by addition of the ssDNA binding protein RPA (Replication Protein A) after Rad51 is pre-nucleated with ssDNA [40][44][45][46][47][48]. The positive effect of RPA was postulated to result from the ability of RPA to remove secondary DNA structures which might impede the formation of continuous, functional Rad51 filaments [47][48][49].
–
–
Rad51 also binds ATP and hydrolases it in a DNA-dependent manner. The ATP binding is required for Rad51 activities [40][44][45][50][51]. Rad51 mutants deficient in nucleotide binding are catalytically dead and cannot form extended nucleofilaments, while mutants that bind ATP but are incapable of hydrolysing it can perform the strand exchange reaction in vitro and partially complement RAD51 deletion in vivo when overexpressed [52][53]. This demonstrates that only ATP binding is required for the essential Rad51 functions but ATP hydrolysis contributes to its full activity inside the cell. However, the role of Rad51 ATP hydrolysis is not completely understood. Possibly, it is important for the disassembly of the filament and recycling of Rad51 pool as S. cerevisiae Rad51, just like its bacterial orthologue RecA, shows decreased affinity for DNA when bound to ADP instead of ATP [54][55][56]. Consistent with this hypothesis, human RAD51 bound to DNA shows lower subunit turnover when ATP hydrolysis is prevented [37][57]. Alternatively, the ATP hydrolysis might promote the DNA annealing step during the strand exchange reaction: while the ATP-bound nucleofilament conformation is stiffer and more suitable for the initial step of separating donor DNA strands, the ADP-bound human RAD51 might promote annealing of the invading and donor strands [58]. Interestingly, in vitro ATP hydrolysis by Rad51 is much slower than that by RecA. When bound to ssDNA, human and yeast orthologues hydrolyse 0.16 and 0.7 ATPs per minute per subunit respectively while RecA turns over 25-30 ATP molecules during that time [40][44][54][59]. Therefore, the modulation of the ATPase activity of Rad51 in vivo by other proteins might have an important function in regulating Rad51 activity.
POSITIVE REGULATORS OF RAD51 NUCLEOPROTEIN FILAMENT FORMATION
As mentioned above, DSB repair by GC and BIR requires a formation of Rad51 filament on resected DNA. However, resected DNA is much more readily coated by the ssDNA binding protein RPA, which is more abundant inside the cells and has a higher affinity for ssDNA than Rad51 [60]. As mentioned earlier, RPA stimulates Rad51-catalysed strand exchange in vitro when it is added after Rad51 has been assembled on ssDNA. However, if RPA is incubated with ssDNA before the Rad51 addition or both proteins are introduced into the reaction simultaneously, Rad51 filament formation and the subsequent strand exchange reaction are largely inhibited [46][48][61][62][63]. This indicates that Rad51 and RPA compete for binding to ssDNA and that Rad51 cannot efficiently replace RPA bound to ssDNA. However, the RPA/Rad51 protein exchange can be facilitated by so called mediator proteins described below.
–
Rad52 and BRCA2
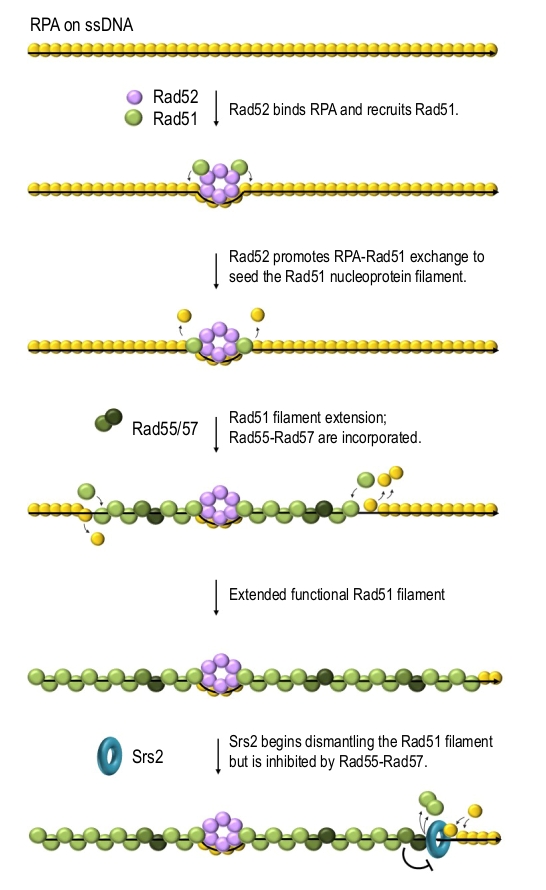
Rad52 is a mediator which promotes Rad51 filament formation on RPA-coated ssDNA [46][48][62][63]. Rad52 can physically interact with DNA, RPA and Rad51 and it has been postulated to promote the formation of Rad51 filaments by both recruiting Rad51 and helping it to replace RPA on ssDNA [46][64][65][66]. Rad52 binds and stabilises RPA on ssDNA while promoting RPA replacement by Rad51 on the Rad52-surrounding DNA, thereby nucleating Rad51 filaments (Figure 3) [67]. The N-terminus of Rad51 has been demonstrated to interact with the DNA binding domain of RPA and possibly promote RPA dissociation [68]. This might allow the nucleated Rad51 to help free Rad51 monomers to compete for DNA binding, thereby extending the filament. This explanation is consistent with the recent in vitro reconstruction studies suggesting that RPA mainly inhibits the nucleation but not the elongation of Rad51 filaments [37][67]. Rad52 also stabilises Rad51 filament via protein-protein interactions [69]. S. cerevisiae cells lacking Rad52 are deficient in DSB repair by HR and cannot form detectable DNA damage-induced Rad51 foci suggesting that Rad51 filament assembly requires Rad52 in vivo [70][71][72].
–
–
Rad52 orthologues are also present in vertebrates. However, RAD52 deficient mice, chicken and human cells show only a slight defect in HR and remain resistant to DSB inducing factors [73][74][75]. This is due to RAD52 playing a secondary role in recombination in mammals while another protein, BRCA2, mediates RAD51 filament formation [76][77]. Like S. cerevisiae Rad52, BRCA2 enables RAD51 to perform the strand exchange reaction with RPA-coated ssDNA substrate in vitro [77]. BRCA2 physically interacts with RAD51, RPA and DNA [78][79]. BRCA2 has been proposed to stabilise RAD51 filaments in vitro by blocking ATP hydro-lysis by RAD51 [77][80]. In addition, one of the BRCA2-interacting proteins, BRCCIPβ, also interacts with RAD51 and induces a conformational change which facilitates the normally slow release of ADP from RAD51, thereby promoting nucleotide exchange and reversal of RAD51 to the active ATP-bound state [81][82]. Another HR accessory factor, HOP2-MND1, can induce RAD51 conformational changes which enhance its ability to bind nucleotides and modulate the ssDNA-binding ability to promote the strand exchange reaction [83].
–
Interestingly, human RAD52 cannot substitute for BRCA2 in nucleating RAD51 on RPA-coated ssDNA in vitro but RAD52 depletion in BRCA2 deficient cells results in synthetic lethality accompanied by a further decrease in HR and a reduction in RAD51 foci induced by ionising radiation [77][84]. This implies that although BRCA2 is the main RAD51 mediator in human cells, RAD52 activity in promoting the assembly of the RAD51 filament is significant enough to keep BRCA2-deficient cells viable.
–
Rad51 paralogues
Yeast Rad51 paralogues Rad55 and Rad57 are two other key mediators. They form a stable heterodimer which physically interacts with Rad51 and can alleviate the RPA-dependent inhibition of Rad51-catalysed strand exchange reaction in vitro [30]. The phenotype of the rad55 rad57 double mutant is identical to those of individual deletions [85]. Mutants lacking Rad55 are sensitive to DNA-damaging agents, deficient in DSB repair by HR and show impaired localisation of Rad51 to an unrepairable DSB suggesting that Rad51 filament assembly is defective in the absence of Rad55-Rad57 [70][86]. The rad51-I345T mutation results in a Rad51 protein with increased affinity for DNA and the ability to overcome the inhibitory effect of RPA during the strand exchange reaction in vitro. rad51-I345T also partially suppresses the DNA damage sensitivity of rad55Δ and rad57Δ cells but it cannot compensate for the loss of the Rad52 mediator activity [85]. Rad55-Rad57 works downstream of the Rad52-dependent Rad51 recruitment to ssDNA and possibly stabilises Rad51 filament through protein-protein interactions, which might maximise the probability that a nucleation event will result in a successful assembly of a functional Rad51 nucleofilament. Rad55-Rad57 has been also proposed to enhance the stability of Rad51 filaments by antagonising Srs2 helicase (described below) [29] which dislodges Rad51 from ssDNA (Figure 3) [87][88]. Consistently, Rad51-I345T which partially compensates for the loss of Rad55-Rad57 is harder for Srs2 to strip from ssDNA in vitro [89] and the DNA damage sensitivity of cells lacking the Rad55-Rad57 complex can be suppressed by a deletion of SRS2 [87][90][91].
–
Five Rad51 paralogues have been described in vertebrates: RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3 [27][28]. They all function in the same pathway as BRCA2 and the depletion of any one of them results in a decreased efficiency of HR in human cells [92]. Phylogenetic studies suggest that XRCC2 and RAD51D are yeast Rad55 and Rad57 orthologues respectively, although some studies place Rad57 most closely related to XRCC3 [28][93][94][95]. The Rad51 paralogs form two stable complexes: one is called BCDX2 and consists of RAD51B, RAD51C, RAD51D, and XRCC2 while the other one – CX3 – is made of RAD51C and XRCC3 [96]. The DNA damage induced formation of RAD51 foci is decreased in human cells depleted of RAD51D, RAD51C and XRCC2 but not XRCC3 suggesting that BCDX2 complex is required for efficient Rad51 filament formation, while CX3 is dispensable for this process [92][97]. In agreement with this, the CX3 complex has been shown to act downstream of Rad51 filament assembly and participate in the resolution of Holliday junctions [98].
–
Shu complex
Another factor that participates in Rad51 filament assembly is the Shu complex. In S. cerevisiae, it consists of the Rad51 paralogues Csm2 and Psy3 which form a heterotetramer along with Shu1 and Shu2 [99][100]. Unlike Rad52 and Rad55-Rad57, the Shu complex is not essential for DSB repair by HR in vivo [99]. However, deletions of the individual genes encoding the Shu complex components do lead to defects in Rad51 foci formation [100]. Csm2 physically interacts with Rad55 and bridges the Shu complex to Rad51 [101]. The Csm2-Psy3 dimer binds DNA and can enhance the Rad51-catalysed strand exchange reaction with RPA-coated substrates in vitro, in a Rad52- and Rad55-Rad57-dependent manner [101]. Although Shu1 and Shu2 are not required for this activity in vitro, shu1Δ results in elevated Srs2-dependent Rad51 filament disassembly in vivo [102]. Furthermore, Shu2 physically interacts with Srs2 suggesting that the Shu complex might promote Rad51 filament assembly by both direct interactions with Rad51 and inhibiting the Srs2-dependent disassembly of the filament [103].
–
A Shu2 orthologue – SWS1 – has been found in humans and shown to interact with another protein, SWSAP1. This interaction mutually stabilises the two proteins and the SWS1-SWSAP1 complex can bind ssDNA. SWSAP1 was also found to physically interact with RAD51, RAD51B, RAD51C, RAD51D and XRCC3 while SWS1 can bind to RAD51D and XRCC3 [104]. Depletion of either SWS1 or SWSAP1 results in decreased formation of DNA damage-induced RAD51 foci suggesting that the function of the Shu complex is conserved from yeast to humans [104][105].
–
Rad54
Other factors, although less significant, have been implicated in promoting Rad51 nucleofilament formation. Rad54/RAD54 is an ATP-dependent translocase which can bind Rad51/RAD51 and DNA simultaneously and plays an important role in synaptic and postsynaptic events. While a catalytically-dead translocase is deficient in DNA strand invasion, it is fully functional in stabilising Rad51 filaments [106][107][108]. When Rad51 is assembled on dsDNA, Rad54 can strip it in an ATP-dependent manner [109][110]. Therefore Rad54 promotes Rad51 filament formation on ssDNA by both stabilising Rad51 binding to ssDNA and inhibiting its association with dsDNA. However, cell cycle dependent phosphorylation of Rad54 can convert it into a negative regulator of Rad51, by enabling a Rad54-dependent removal of Rad51 at HR loci in G2 [111].
–
Swi5-Srf1 (Sae3-Mei5)
Sae3 and Mei5 are meiosis-specific mediators of Rad51 and Dmc1 filament formation in S. cerevisiae [112]. However, their conserved orthologues in other organisms, including S. pombe, mice and humans (Swi5/SWI5 and Sfr1/SFR1 respectively), participate in the assembly of mitotic Rad51/RAD51 filament [113][114][115]. Swi5/SWI5 and Sfr1/SFR1 form a stable complex with a 1:1 stoichiometry and can directly interact with Rad51/RAD51 [114][115][116].
–
S. pombe Swi5-Sfr1 stimulates the in vitro Rad51-dependent strand exchange reaction [117]. Furthermore, the formation of Rad51/RAD51 foci after exposure to ionising radiation is decreased in both sfr1Δ S. pombe and SRF1 depleted human cells, pointing towards a defect in the assembly of Rad51 [113][114]. S. pombe Swi5 and Sfr1 proteins form an elongated structure in vitro which fits in the helical groove of the presynaptic filament, thereby suggesting a hypothetical mechanism for Swi5-Sfr1 action during the assembly of Rad51 [116].
–
INO80
It is important to note that Rad51 assembles on DNA in a context of chromatin. Not surprisingly, an evolutionally conserved nucleosome remodelling complex INO80 has been linked to the formation of the presynaptic filament [118][119][120][121]. Disruption of the INO80 remodelling complex in budding yeast and human cells results in a decreased efficiency of HR. Rad51 accumulation on resected DNA is also reduced pointing to a defect in the formation of the nucleofilament. This phenotype can be largely suppressed by a removal of the H2A.Z histone variant suggesting that H2A.Z has an inhibitory effect on the assembly of Rad51 and that INO80 complex is required to remove it from damaged chromatin [118][119][122][123]. However, the exact mechanism of how H2A.Z inhibits Rad51 filament formation is unknown. Although DNA resection is also affected in cells lacking INO80 [118][119], the H2A.Z removal suppresses the defect of Rad51 filament assembly but it does not compensate for the resection defect [119]. As HR efficiency is also largely restored in cells lacking both INO80 and H2A.Z, the main function of INO80 in HR is likely to be the facilitation of Rad51 nucleofilament formation [118][119].
NEGATIVE REGULATION OF RAD51-DNA BINDING
Rad51 removal from undamaged dsDNA
As mentioned above, Rad51 can bind both ssDNA and dsDNA in vitro [24][39]. While Rad51 binding to ssDNA is essential for the strand exchange reaction in vitro, pre-coating dsDNA with Rad51 inhibits the formation of the product [45][109]. In vivo, Rad51 cellular pools are limited and Rad51 binding to dsDNA depletes the pool of free monomers leaving fewer of them available for the repair [70]. Thus, Rad51 binding to undamaged chromatin can impede Rad51-dependent recombination and may result in genome instability and chromosome loss [110][124].
–
In S. cerevisiae, non-damage-associated DNA binding of Rad51 is cytologically undetectable as it is actively antagonised by three SWI2/SNF2 family DNA translocases – Rad54, Rdh54 and Uls1 [124]. Both Rad54 and Rdh54 remove Rad51 from dsDNA in an ATP-dependent manner in vitro [109][125]. This activity requires the N-terminal parts of Rad54 and Rdh54 which have been shown to interact with Rad51 but the exact mechanism of Rad51 displacement is unknown [126][127]. Rad54 has been further demonstrated to enable the strand exchange reaction in vitro, even when both ssDNA and the donor dsDNA are covered with Rad51 [109].
–
Rdh54 might be the main player in the removal of Rad51 from undamaged chromatin as the lack of this protein but not the other two translocases results in accumulation of spontaneous non-damage-associated Rad51 foci [124]. However, deletion of all the three translocase-coding genes results in a more severe phenotype suggesting that Rad54 and Uls1 can partially substitute for Rdh54 [124][128]. Consistent with the in vitro evidence, inactivation of the ATPase activity in any of the three proteins results in the same phenotype as in the corresponding deletion mutants, further highlighting the need for the Rdh54 and Rad54 translocase activities in the removal of Rad51 from dsDNA; Uls1 is likely to operate in a similar way [124].
–
Rad54 homologs are well-conserved among eukaryotes, with human cells containing two known proteins – RAD54 and RAD54B [129]. Like its yeast counterpart, human RAD54 can remove RAD51 from dsDNA in vitro [110]. Simultaneous depletion of both RAD54 and RAD54B results in accumulation of RAD51 on chromatin in human tumour cells [110]. The fact that both proteins need to be depleted to reveal the phenotype suggests that they are redundant and that RAD54B also may have the ability to remove RAD51 from dsDNA. This suggests that the function of the discussed SWI2/SNF2 translocases in the regulation of the RAD51 cellular pool might be conserved from yeasts to humans.
–
Suppression of HR at replication forks via inhibition of Rad51 filament formation
DNA synthesis in eukaryotes strongly depends on PCNA (Proliferating Cell Nuclear Antigen), a homotrimeric protein which forms a ring around the DNA and acts as a tool belt holding different components of the replication machinery: polymerases, ligases, nucleases, helicases, etc. [130]. Some of these enzymes, such as DNA polymerases, are associated with PCNA at the fork almost all the time while others can be recruited as needs for their activity at specific loci arise. For example, Pif1 family helicases are recruited to hard-to-replicated loci [131]. Replication fork barriers (tightly bound proteins, G-rich DNA regions, highly expressed genes, etc) cause replication fork pausing which often leads to accumulation of ssDNA, followed by recruitment of Rad52 and Rad51, thereby creating an opportunity for the DNA at the fork to be involved in unwanted HR. In budding yeast, these potentially mutagenic events are prevented by the Srs2 helicase which has been shown to disassemble the Rad51 nucleoprotein filament by dislodging Rad51 from ssDNA [87][88]. Srs2 is recruited to replication forks through a direct binding of the SUMO-interacting motif (SIM) and the PCNA-interacting peptide box in the C-terminus of Srs2 to a SUMOylated PCNA [132][133][134]. Once recruited to replication forks, Srs2 removes Rad51 and prevents potentially deleterious unscheduled HR events that might occur when replication fork progression is slowed down or paused by replication barriers. Disruption of this regulatory mechanism by mutations in SRS2 leads to a hyper-recombination phenotype (increased mitotic recombination) [135][136]. However, when replication forks are stalled due to a damaged template, Rad51 recruitment to the replication fork might be desirable in order to bypass the DNA lesion via the template switching mechanism [137]. In this case, local Srs2 levels might be decreased through targeting the fork-bound Srs2 for proteasomal degradation [138]. In addition, PCNA can be unloaded from stalled replication forks by the Elg1-containing RFC complex, thereby eliminating the PCNA-dependent binding of Srs2 to the fork [138].
–
Recent advances in understanding the role of RAD51 at replication forks in higher eukaryotes suggest an additional layer of regulatory mechanisms modulating RAD51 activities during replication. In human cells, a depletion of either BRCA2 or RAD51 results in under-replication and cell cycle arrest in the subsequent G1 phase [139]. The current understanding of replication considers stalled replication forks as part of normal cell physiology and fork reversal as a mechanism stabilising the forks [18]. RAD51 has been proposed to participate in both fork reversal [140] and protection of these forks from excessive DNA degradation by nucleases [141][142]. A reversed replication fork is a four-way dsDNA junction, with one of the four branches terminating in a one-ended DSB which can serve as an entry point for the break resection machinery. BRCA2-dependent recruitment of RAD51 to a partially resected reversed fork has been proposed to inhibit further DNA degradation [142][143]. On the other hand, RAD51 accumulation at replication forks is counteracted by a newly-identified ssDNA-binding protein RADX which prevents fork collapse due to excessive activity of RAD51 [144]. Interestingly, deleting RADX restores fork protection in BRCA2-deficient cells [144] suggesting that a fine balance between the positive and negative regulation of RAD51 at the forks by BRCA2 and RADX respectively is required for the genome stability maintenance during replication. Rad51 was further demonstrated to physically interact with the primase Polα, possibly by recruiting it to stalled replication forks and promoting their restart [142]. Rad51 also prevents the degradation of nascent DNA at ssDNA gaps which might form behind replication forks due to a damaged DNA template. Normally, these gaps are small and might be undetectable but accumulation of extensive ssDNA tracks behind replication forks has been observed in the absence of Rad51 binding to chromatin in Xenopus egg extracts. The accumulation of these ssDNA gaps may be suppressed by the inhibition of the Mre11 nuclease activity, further supporting the role of Rad51 in the protection of nascent DNA against the degradation by nucleases [141].
–
The role of yeast Rad51 in fork protection is less understood but some close similarities to the findings in higher eukaryotes have been found: RAD51 deletion in S. cerevisiae leads to accumulation of ssDNA gaps at forks and behind them. Rad51 and Rad52 localise to the forks during replication and are required for post-replicative DNA repair via HR [145]. Therefore, the replication-associated functions of Rad51 might be conserved in eukaryotes.
–
Disassembly of Rad51 nucleoprotein filament during DSB repair
On one hand, disassembly of Rad51 presynaptic filaments during DSB repair may play a role in limiting excessive recombination events. On the other hand, the same activity plays pro-recombination role as it is required at the late stages of repair to ‘clean up’ postsynaptic DNA in order to enable recruitment of PCNA and the rest of the replication machinery to re-synthesize resected DNA [17]. The Srs2 helicase has now been implicated in both functions and therefore Srs2-dependent disassembly of Rad51 filament previously considered inhibitory to HR, also has a pro-recombination role as discussed below.
–
PCNA is recruited to DNA by Replication Factor C (RFC) which recognises primer-template junctions and loads PCNA on dsDNA [146][147][148]. The RFC-PCNA complex can bind primer-template DNA junctions and in general has a significant affinity to ssDNA in vitro. The latter can be inhibited by the addition of RPA, which enhances the specificity of PCNA loading to the junctions [148][149][150]. RPA physically interacts with RFC and stimulates PCNA loading onto DNA in vitro [17][151][152][153]. In contrast, Rad51 inhibits PCNA loading but this inhibition can be suppressed by either increasing the concentrations of RPA, which competes with Rad51 for ssDNA binding, or by addition of the Srs2 helicase which disassembles Rad51 filaments, thereby promoting RPA binding to DNA and the consequential PCNA recruitment via RPA-RFC interactions [17][88][152].
–
Srs2 can remove RPA, Rad52 and Rad51 from ssDNA in vitro [55][87][88][89][154]. Srs2 can physically interact with Rad51 and although S. cerevisiae Srs2 is capable of removing human RAD51 in vitro, the efficient clearance of ssDNA from Rad51 depends on species-specific interactions between Srs2 and Rad51, as well as ATP hydrolysis by Rad51 and Srs2 translocase activity [55][89]. It has been suggested that Srs2 allosterically activates ATP hydrolysis in Rad51 monomers, thereby decreasing their affinity for DNA. Srs2 translocation is postulated to be important for the processivity and positioning of the helicase, which might be required to make appropriate contacts with the successive Rad51 monomers [55]. In addition, a tandem assembly of Srs2 monomers appears to be important for the efficient disassembly of Rad51 filaments [89]. On the other hand, the rate of Rad51 stripping is also influenced by the strength of the Rad51-ssDNA interaction: the amino acid substitutions which increase the stability of Rad51 on ssDNA due to either increased affinity for ssDNA or inability to hydrolyse ATP both slow down the rates of Rad51 removal by Srs2 [89].
–
Importantly, Srs2 has been proposed to promote SDSA by disrupting D-loops in a manner that requires its ATPase activity, Rad51-interacting domain, SIM and PCNA-interacting peptide box [136][155][156][157]. SRS2 deletion leads to an elevated frequency of crossovers during the repair of an induced DSB [155].
–
It is not known in detail how Srs2 is brought to DNA repair sites but its localisation is independent of the SIM which is required for Srs2 recruitment to stalled replication forks via interaction with SUMOylated PCNA [133]. The Srs2-ΔC (1-860) mutant protein lacking the Rad51-interacting region of Srs2 has a greatly impaired Rad51 clearance activity in vitro, mainly due to a decreased loading of mutant Srs2 on Rad51-ssDNA filaments [55][89]. This suggests that the Rad51-interacting region of Srs2 promotes Srs2 association with the presynaptic filament [89]. However, Srs2 has been shown to localise to repair sites even in the absence of Rad51 [133]. Furthermore, the srs2-ΔC (1-860) allele can suppress the DNA re-synthesis defects arising from the lack of Rad51 removal in srs2Δ cells [17]. These observations suggest that there might be a Rad51-independent way to recruit Srs2 to repair sites in vivo. Consistently, it has been recently demonstrated that Srs2 can directly bind heteroduplex DNA joints in vitro [89].
–
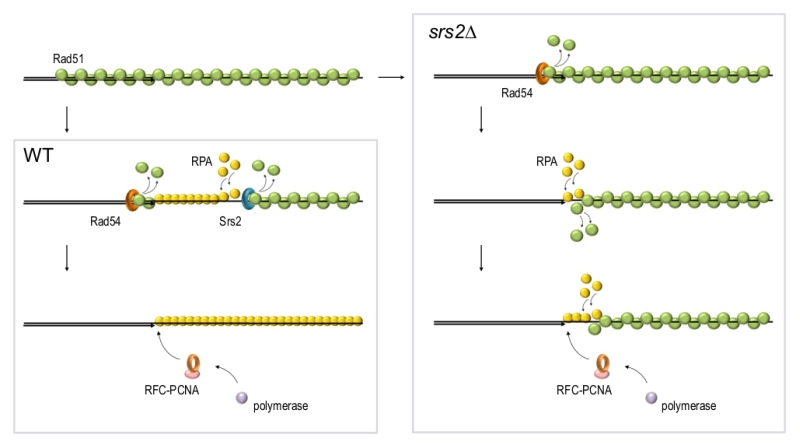
Inability to disassemble Rad51 filaments at the repair sites in srs2Δ cells leads to accumulation of ssDNA gaps, persistence of the DNA damage signalling and failure to complete DNA repair [17][158][159]. These ssDNA gaps stem from a defect in re-synthesis of resected DNA which is likely required to terminate DNA resection. Although the re-synthesis is impaired in srs2Δ, the reconstitution of dsDNA during DSB repair can be observed in srs2Δ, albeit at a slower rate [17]. This could be explained by a sufficiently high stochastic exchange between Rad51 and RPA on ssDNA. Alternatively, there might be other yet unidentified proteins involved in Rad51 removal during the late stages of repair. Our recent experiments indicate that Rad54 might be such a protein as the loss of both Srs2 and Rad54 almost completely blocks re-synthesis of resected DNA during DSB repair (Andriuskevicius and Makovets, unpublished results). Our current hypothesis is that while Srs2 removes Rad51 from ssDNA, Rad54 might be disrupting the extension of the Rad51 filament at the neighbouring dsDNA region (Figure 4). In the absence of Srs2, the Rad54-dependent removal of Rad51 from dsDNA might extend into the ssDNA. Alternatively, stripping of Rad51 from dsDNA by Rad54 might promote stochastic dissociation of Rad51 from the ssDNA at the junction and vice versa: dislodging of Rad51 by Srs2 from ssDNA may destabilise Rad51 bound to the dsDNA at the junction (Figure 4). This hypothesis is consistent with the observations that stochastic Rad51 dissociation from DNA is higher for the monomers at the end of the filament than for the internal ones [37][160]. The dissociation of Rad51 from the dsDNA at the junction might be required not only for the recruitment of PCNA but also to make the 3’end accessible for DNA polymerases in order to initiate DNA polymerisation, both during re-synthesis of resected DNA and when extending the invaded DNA strand in a D-loop [161]. This might explain why the post-invasion DNA synthesis during BIR in srs2Δ is also affected [17]. To summarise, in yeast Rad54 and Srs2 might be complementing each other in disassembly of Rad51 filaments while being partially redundant in Rad51 removal from the ssDNA-dsDNA junction due to the intrinsic features of the Rad51 filament. Indirect evidence suggest that RAD54 might have a similar function in vertebrates [111][162].
–
–
A clear homolog of Srs2 has not been found in higher eukaryotes but other proteins have the ability to destabilise RAD51 nucleofilaments. G2-induced phosphorylation of Rad54 in Xenopus enables it to remove Rad51 from sites of HR [111]. Human helicases FBH1, BLM, RECQL5 and FANCJ have been demonstrated to dislodge RAD51 from ssDNA in an ATP-dependent manner in vitro [163][164][165][166]. Another protein called PARI also has the ability to remove RAD51 from ssDNA in vitro. Although PARI does not have an ATPase activity, the PARI-mediated disassembly of the nucleofilament is dependent on ATP hydrolysis by RAD51 suggesting that PARI, like Srs2, can stimulate RAD51 ATPase activity [167]. This also supports the idea that ATP hydrolysis destabilises human RAD51 filament. Similarly to Srs2, PARI also functions at replication forks where it is recruited via PCNA and PCNA SUMOylation facilitates this recruitment [167]. Mammalian cells lacking FBH1, BLM, RECQL5 or PARI exhibit hyper-recombination phenotypes suggesting the relevance of these proteins to the negative control of recombination in vivo [165][167][168][169][170].
–
Human cells might also be able to remove RAD51 from DNA at DNA damage sites via ubiquitination [162]. The RFWD3 E3 ubiquitin ligase can physically interact with RAD51 and polyubiquitinate it both in vitro and in vivo. The RFWD3-dependent ubiquitination of RAD51 results in its degradation by proteasome. Cells depleted of RFWD3 or expressing an ubiquitination-deficient RAD51 variant show decreased turnover of RAD51 at repair sites, suggesting that the ubiquitination might destabilise RAD51 in the filament by targeting it for degradation. Although the exact stage of HR involving RFWD3-dependent regulation is unknown, it is proposed to function after the formation of RAD51 nucleofilament, and perhaps, even downstream of postsynapsis. If RFWD3 were destabilising RAD51 at the presynaptic stage and working as an anti-recombinase, the HR frequency would have been increased when RAD51 ubiquitination was prevented. However, the opposite has been observed in cells expressing ubiquitination-defective RAD51 mutant protein, suggesting that RFWD3-assisted RAD51 removal is important for the progression of HR at the later stages, possibly during the postsynaptic strand extension [162]. Interestingly, decreased RAD54 chromatin loading was observed in RFWD3 deficient human cells after DNA damage, suggesting that RAD51 ubiquitination and RAD54 loading might be functionally linked [162].
–
It is important to emphasise that Rad51 filament is formed on resected DNA breaks independently of whether the repair will proceed by HR or by some other mechanism, for example, SSA or DNTA. Whichever route the repair takes, eventually the resected DNA is to be re-synthesised and converted back to dsDNA, in order to complete the repair and switch off the DNA damage signalling. Although Rad51 is not required for DSB repair via either SSA or DNTA, the Srs2-dependent activity on Rad51 is necessary for these mechanisms to be efficient [17]. The results on DNTA are particularly interesting: they suggest that DSB healing by telomerase normally happens on resected DNA ends, although DNTA is dramatically increased in mutants with deficient resection [171][172]. The evidence for DNTA and BIR raises a possibility that any DNA repair mechanisms that involve generation of ssDNA might be affected by unwanted Rad51 filament formation if the ssDNA is persistent long enough for the filament to assemble. Therefore, Rad51 removal by Srs2 or other means might be required not only during DSB repair but have a broader function in DNA repair.
REGULATION OF RAD51 FILAMENT DYNAMICS THROUGH POST-TRANSLATIONAL MODIFICATIONS
Rad51 functions are regulated through post-translational modifications (PTMs) of not only Rad51 but also of the positive and negative regulators of Rad51 nucleoprotein filament assembly. The purpose of most of these modifications falls into one of the three categories: i) cell cycle dependent restrictions on the activity of a factor; ii) upregulation of DNA repair in cells with DNA damage; and iii) regulation of protein turnover at repair sites. Homology-based repair is limited to S-G2, mostly due to the cell cycle dependent phosphorylation required to activate the resection endonucleases upon entry in the S phase. This might explain why Rad52 localisation to DNA depends on the CDK1 activity [173][174] although CDK1-dependent phosphorylation of Rad52 has been detected in high-throughput experiments [175] and therefore Rad52 might be also directly regulated by CDK1. SUMOylation is involved in regulation of Rad52 too: it leads to stabilisation of the protein in vivo [176] while decreasing its affinity to ssDNA [177]. In yeast, both Rad51 and Rad55 are phosphorylated in response to DNA damage and these PTMs are required for cell survival upon DNA damage induction [178][179][180]. Activation of the DNA damage response in yeast as well as mammals leads to RPA SUMOylation [181][182][183] which increases its interaction with RAD51, possibly through the recently identified SIM within RAD51 [184]. Both RAD51 and BRCA2 are deubiquitinated in response to DNA damage [185][186]: deubiquitination of BRCA2 leads to its stabilisation [186] while deubiquitinated RAD51 increases its binding to BRCA2 [185]. In turn, RAD51 ubiquitination might be important for RAD51 removal at the later stages of repair possibly by RAD54 [162], as discussed above. The removal of Rad51 from repair foci in G2 in Xenopus is promoted by cell cycle specific phosphorylation of Rad54 [111].
–
The role of multiple PTMs in regulation of Srs2 is well-documented. CDK1-dependent phosphorylation at the C-terminus promotes the interaction of Srs2 with Mre11 [187], counteracts its SUMOylation and directs Srs2 to unwinding D-loops, thereby promoting DSB repair by SDSA [188]. Although this phosphorylation controls the localisation of the Srs2 to D-loops, neither the elimination of the phosphorylation nor SRS2 deletion has an effect on Rad51 presence at the site of strand invasion, suggesting that Srs2 acts downstream of the invasion step [188]. The C-terminus of Srs2 also plays an important role in regulation of its function at replication forks. As mentioned above, the C-terminus of Srs2 contains a SIM which is required for Srs2 interaction with PCNA at replication forks. However, the C-terminus also contains SUMOylation sites and Srs2 SUMOylation inhibits its interaction with PCNA [189]. Therefore, PTMs of Srs2 regulate both its anti-recombinase role at replication forks and its pro-recombination function in SDSA. The C-terminus is not required for the role of Srs2 in Rad51 removal during re-synthesis of resected DNA [17]. However, the shortest functional C-terminal truncation Srs2 (1-860) retains two out of the seven identified CDK1 sites which might be important for this function. In summary, regulation of HR in general and Rad51 filament assembly in particular through PTM provides an additional layer of mechanisms to boost Rad51 activities in response to DNA damage, fine-tune them depending on the type of DNA damage and cell cycle stage as well as enables cells to regulate Rad51 and its interaction with DNA and other proteins as the repair progresses.
CONCLUSION
The importance of recombinases in genome stability and diversity has been appreciated since the early days of molecular biology. Classical biochemistry and crystallography followed by more recently emerged single-molecule analysis and cryo-electron microscopy have provided molecular insights into their structure and function at the molecular level. We have also learned that numerous accessory proteins are required for a recombinase to operate in vivo; a whole set of enzymes built around it comprises HR machinery. The complexity of regulatory mechanisms governing this machinery has been becoming more and more obvious with the increasing number of discoveries of PTMs which modulate the components of HR machinery and their regulators. Rad51 filament formation is one of the most critical steps in HR and its regulation, both positive (filament assembly) and negative (disassembly) are required for efficient DNA repair. The newly-emerging role of recombinases at stalled replication forks adds another angle to understanding biological significance of their regulation. The sophistication of this regulatory network, based on the multitude of inputs and connections to other cellular pathways, increases from prokaryotes to yeast to higher eukaryotes, thereby underlying the importance of the fine-tuning of recombinase activities for cell survival and adaptive evolution of species.
References
- P.J. McKinnon, and K.W. Caldecott, "DNA Strand Break Repair and Human Genetic Disease", Annual Review of Genomics and Human Genetics, vol. 8, pp. 37-55, 2007. http://dx.doi.org/10.1146/annurev.genom.7.080505.115648
- K. Rodgers, and M. McVey, "Error‐Prone Repair of DNA Double‐Strand Breaks", Journal of Cellular Physiology, vol. 231, pp. 15-24, 2015. http://dx.doi.org/10.1002/jcp.25053
- K.M. Kramer, J.A. Brock, K. Bloom, J.K. Moore, and J.E. Haber, "Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events.", Molecular and Cellular Biology, vol. 14, pp. 1293-1301, 1994. http://dx.doi.org/10.1128/mcb.14.2.1293
- V. Gorbunova, "Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions", Nucleic Acids Research, vol. 25, pp. 4650-4657, 1997. http://dx.doi.org/10.1093/nar/25.22.4650
- A. Smogorzewska, J. Karlseder, H. Holtgreve-Grez, A. Jauch, and T. de Lange, "DNA Ligase IV-Dependent NHEJ of Deprotected Mammalian Telomeres in G1 and G2", Current Biology, vol. 12, pp. 1635-1644, 2002. http://dx.doi.org/10.1016/s0960-9822(02)01179-x
- H. Ghezraoui, M. Piganeau, B. Renouf, J. Renaud, A. Sallmyr, B. Ruis, S. Oh, A.E. Tomkinson, E. Hendrickson, C. Giovannangeli, M. Jasin, and E. Brunet, "Chromosomal Translocations in Human Cells Are Generated by Canonical Nonhomologous End-Joining", Molecular Cell, vol. 55, pp. 829-842, 2014. http://dx.doi.org/10.1016/j.molcel.2014.08.002
- J. Zámborszky, B. Szikriszt, J.Z. Gervai, O. Pipek, �. Póti, M. Krzystanek, D. Ribli, J.M. Szalai-Gindl, I. Csabai, Z. Szallasi, C. Swanton, A.L. Richardson, and D. Szüts, "Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions", Oncogene, vol. 36, pp. 746-755, 2016. http://dx.doi.org/10.1038/onc.2016.243
- S.P. Jackson, "Sensing and repairing DNA double-strand breaks", Carcinogenesis, vol. 23, pp. 687-696, 2002. http://dx.doi.org/10.1093/carcin/23.5.687
- L. Bee, S. Fabris, R. Cherubini, M. Mognato, and L. Celotti, "The Efficiency of Homologous Recombination and Non-Homologous End Joining Systems in Repairing Double-Strand Breaks during Cell Cycle Progression", PLoS ONE, vol. 8, pp. e69061, 2013. http://dx.doi.org/10.1371/journal.pone.0069061
- C. Ribeyre, and D. Shore, "Regulation of telomere addition at DNA double-strand breaks", Chromosoma, vol. 122, pp. 159-173, 2013. http://dx.doi.org/10.1007/s00412-013-0404-2
- J. Chen, D.N. Cooper, N. Chuzhanova, C. Férec, and G.P. Patrinos, "Gene conversion: mechanisms, evolution and human disease", Nature Reviews Genetics, vol. 8, pp. 762-775, 2007. http://dx.doi.org/10.1038/nrg2193
- A. Malkova, and G. Ira, "Break-induced replication: functions and molecular mechanism", Current Opinion in Genetics & Development, vol. 23, pp. 271-279, 2013. http://dx.doi.org/10.1016/j.gde.2013.05.007
- R. Bhargava, D.O. Onyango, and J.M. Stark, "Regulation of Single-Strand Annealing and its Role in Genome Maintenance", Trends in Genetics, vol. 32, pp. 566-575, 2016. http://dx.doi.org/10.1016/j.tig.2016.06.007
- L.S. Symington, "Mechanism and regulation of DNA end resection in eukaryotes", Critical Reviews in Biochemistry and Molecular Biology, vol. 51, pp. 195-212, 2016. http://dx.doi.org/10.3109/10409238.2016.1172552
- N. Saini, S. Ramakrishnan, R. Elango, S. Ayyar, Y. Zhang, A. Deem, G. Ira, J.E. Haber, K.S. Lobachev, and A. Malkova, "Migrating bubble during break-induced replication drives conservative DNA synthesis", Nature, vol. 502, pp. 389-392, 2013. http://dx.doi.org/10.1038/nature12584
- J.M. Stark, and M. Jasin, "Extensive Loss of Heterozygosity Is Suppressed during Homologous Repair of Chromosomal Breaks", Molecular and Cellular Biology, vol. 23, pp. 733-743, 2003. http://dx.doi.org/10.1128/mcb.23.2.733-743.2003
-
Y. Vasianovich, V. Altmannova, O. Kotenko, M.D. Newton, L. Krejci, and S. Makovets, "Unloading of homologous recombination factors is required for restoring double‐stranded
DNA at damage repair loci", The EMBO Journal, vol. 36, pp. 213-231, 2017. http://dx.doi.org/10.15252/embj.201694628 -
A.M. Kolinjivadi, V. Sannino, A. de Antoni, H. Técher, G. Baldi, and V. Costanzo, "Moonlighting at replication forks – a new life for homologous recombination proteins
BRCA 1,BRCA 2 andRAD 51", FEBS Letters, vol. 591, pp. 1083-1100, 2017. http://dx.doi.org/10.1002/1873-3468.12556 - J. Game, and R. Mortimer, "A genetic study of X-ray sensitive mutants in yeast", Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, vol. 24, pp. 281-292, 1974. http://dx.doi.org/10.1016/0027-5107(74)90176-6
- S. Nakai, and S. Matsumoto, "Two types of radiation-sensitive mutant in yeast", Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, vol. 4, pp. 129-136, 1967. http://dx.doi.org/10.1016/0027-5107(67)90064-4
- H. HAMA-INABA, and T. SAEKI, "Evidence that a Recombination less Strain, rod 51, of Saccharomyces cerevisiae Lacks the Budding Cell Resistance to γ-rays", Journal of Radiation Research, vol. 16, pp. 37-44, 1975. http://dx.doi.org/10.1269/jrr.16.37
- M.A. Resnick, "The repair of double-strand breaks in DNA: A model involving recombination", Journal of Theoretical Biology, vol. 59, pp. 97-106, 1976. http://dx.doi.org/10.1016/s0022-5193(76)80025-2
- W.J. Jachymczyk, R.C. von Borstel, M.R.A. Mowat, and P.J. Hastings, "Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: The RAD3 system and the RAD51 system", Molecular and General Genetics MGG, vol. 182, pp. 196-205, 1981. http://dx.doi.org/10.1007/bf00269658
- A. Shinohara, H. Ogawa, and T. Ogawa, "Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein", Cell, vol. 69, pp. 457-470, 1992. http://dx.doi.org/10.1016/0092-8674(92)90447-k
- A. Malkova, E.L. Ivanov, and J.E. Haber, "Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication.", Proceedings of the National Academy of Sciences, vol. 93, pp. 7131-7136, 1996. http://dx.doi.org/10.1073/pnas.93.14.7131
- N.Y. Stassen, J.M. Logsdon Jr., G.J. Vora, H.H. Offenberg, J.D. Palmer, and M.E. Zolan, "Isolation and characterization of rad51 orthologs from Coprinus cinereus and Lycopersicon esculentum, and phylogenetic analysis of eukaryotic recA homologs", Current Genetics, vol. 31, pp. 144-157, 1997. http://dx.doi.org/10.1007/s002940050189
- S.V. Chintapalli, G. Bhardwaj, J. Babu, L. Hadjiyianni, Y. Hong, G.K. Todd, C.A. Boosalis, Z. Zhang, X. Zhou, H. Ma, A. Anishkin, D.B. van Rossum, and R.L. Patterson, "Reevaluation of the evolutionary events within recA/RAD51 phylogeny", BMC Genomics, vol. 14, 2013. http://dx.doi.org/10.1186/1471-2164-14-240
- Z. Lin, H. Kong, M. Nei, and H. Ma, "Origins and evolution of the recA / RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer", Proceedings of the National Academy of Sciences, vol. 103, pp. 10328-10333, 2006. http://dx.doi.org/10.1073/pnas.0604232103
- J. Liu, L. Renault, X. Veaute, F. Fabre, H. Stahlberg, and W. Heyer, "Rad51 paralogues Rad55–Rad57 balance the antirecombinase Srs2 in Rad51 filament formation", Nature, vol. 479, pp. 245-248, 2011. http://dx.doi.org/10.1038/nature10522
- P. Sung, "Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase.", Genes & Development, vol. 11, pp. 1111-1121, 1997. http://dx.doi.org/10.1101/gad.11.9.1111
- D. Lim, and P. Hasty, "A Mutation in Mouse rad51 Results in an Early Embryonic Lethal That Is Suppressed by a Mutation in p53", Molecular and Cellular Biology, vol. 16, pp. 7133-7143, 1996. http://dx.doi.org/10.1128/mcb.16.12.7133
- E. Sonoda, "Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death", The EMBO Journal, vol. 17, pp. 598-608, 1998. http://dx.doi.org/10.1093/emboj/17.2.598
- X. Yu, S.A. Jacobs, S.C. West, T. Ogawa, and E.H. Egelman, "Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA", Proceedings of the National Academy of Sciences, vol. 98, pp. 8419-8424, 2001. http://dx.doi.org/10.1073/pnas.111005398
- C. Danilowicz, A. Peacock-Villada, J. Vlassakis, A. Facon, E. Feinstein, N. Kleckner, and M. Prentiss, "The differential extension in dsDNA bound to Rad51 filaments may play important roles in homology recognition and strand exchange", Nucleic Acids Research, vol. 42, pp. 526-533, 2013. http://dx.doi.org/10.1093/nar/gkt867
- J.M. Short, Y. Liu, S. Chen, N. Soni, M.S. Madhusudhan, M.K. Shivji, and A.R. Venkitaraman, "High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy", Nucleic Acids Research, pp. gkw783, 2016. http://dx.doi.org/10.1093/nar/gkw783
- A.B. Conway, T.W. Lynch, Y. Zhang, G.S. Fortin, C.W. Fung, L.S. Symington, and P.A. Rice, "Crystal structure of a Rad51 filament", Nature Structural & Molecular Biology, vol. 11, pp. 791-796, 2004. http://dx.doi.org/10.1038/nsmb795
- C.J. Ma, B. Gibb, Y. Kwon, P. Sung, and E.C. Greene, "Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament", Nucleic Acids Research, vol. 45, pp. 749-761, 2016. http://dx.doi.org/10.1093/nar/gkw1125
- K. McEntee, G.M. Weinstock, and I.R. Lehman, "Binding of the recA protein of Escherichia coli to single- and double-stranded DNA.", The Journal of biological chemistry, 1981. http://www.ncbi.nlm.nih.gov/pubmed/7021553
- E.M. Zaitseva, E.N. Zaitsev, and S.C. Kowalczykowski, "The DNA binding properties of Saccharomyces cerevisiae Rad51 protein.", The Journal of biological chemistry, 1999. http://www.ncbi.nlm.nih.gov/pubmed/9915828
- P. Baumann, F.E. Benson, and S.C. West, "Human Rad51 Protein Promotes ATP-Dependent Homologous Pairing and Strand Transfer Reactions In Vitro", Cell, vol. 87, pp. 757-766, 1996. http://dx.doi.org/10.1016/s0092-8674(00)81394-x
- T.K. Prasad, C.C. Yeykal, and E.C. Greene, "Visualizing the Assembly of Human Rad51 Filaments on Double-stranded DNA", Journal of Molecular Biology, vol. 363, pp. 713-728, 2006. http://dx.doi.org/10.1016/j.jmb.2006.08.046
- X. Zhang, V.E. Galkin, X. Yu, E.H. Egelman, and W. Heyer, "Loop 2 in Saccharomyces cerevisiae Rad51 protein regulates filament formation and ATPase activity", Nucleic Acids Research, vol. 37, pp. 158-171, 2008. http://dx.doi.org/10.1093/nar/gkn914
- Y. Matsuo, I. Sakane, Y. Takizawa, M. Takahashi, and H. Kurumizaka, "Roles of the human Rad51 L1 and L2 loops in DNA binding", The FEBS Journal, vol. 273, pp. 3148-3159, 2006. http://dx.doi.org/10.1111/j.1742-4658.2006.05323.x
- P. Sung, "Catalysis of ATP-Dependent Homologous DNA Pairing and Strand Exchange by Yeast RAD51 Protein", Science, vol. 265, pp. 1241-1243, 1994. http://dx.doi.org/10.1126/science.8066464
- P. Sung, and D.L. Robberson, "DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA", Cell, vol. 82, pp. 453-461, 1995. http://dx.doi.org/10.1016/0092-8674(95)90434-4
- P. Sung, "Function of Yeast Rad52 Protein as a Mediator between Replication Protein A and the Rad51 Recombinase", Journal of Biological Chemistry, vol. 272, pp. 28194-28197, 1997. http://dx.doi.org/10.1074/jbc.272.45.28194
- T. Sugiyama, E.M. Zaitseva, and S.C. Kowalczykowski, "A Single-stranded DNA-binding Protein Is Needed for Efficient Presynaptic Complex Formation by the Saccharomyces cerevisiae Rad51 Protein", Journal of Biological Chemistry, vol. 272, pp. 7940-7945, 1997. http://dx.doi.org/10.1074/jbc.272.12.7940
- B. Song, and P. Sung, "Functional Interactions among Yeast Rad51 Recombinase, Rad52 Mediator, and Replication Protein A in DNA Strand Exchange", Journal of Biological Chemistry, vol. 275, pp. 15895-15904, 2000. http://dx.doi.org/10.1074/jbc.M910244199
- A.L. Eggler, R.B. Inman, and M.M. Cox, "The Rad51-dependent Pairing of Long DNA Substrates Is Stabilized by Replication Protein A", Journal of Biological Chemistry, vol. 277, pp. 39280-39288, 2002. http://dx.doi.org/10.1074/jbc.M204328200
- T. Ogawa, X. Yu, A. Shinohara, and E.H. Egelman, "Similarity of the Yeast RAD51 Filament to the Bacterial RecA Filament", Science, vol. 259, pp. 1896-1899, 1993. http://dx.doi.org/10.1126/science.8456314
- F.E. Benson, A. Stasiak, and S.C. West, "Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA.", The EMBO journal, 1994. http://www.ncbi.nlm.nih.gov/pubmed/7988572
- C. Morrison, A. Shinohara, E. Sonoda, Y. Yamaguchi-Iwai, M. Takata, R.R. Weichselbaum, and S. Takeda, "The Essential Functions of Human Rad51 Are Independent of ATP Hydrolysis", Molecular and Cellular Biology, vol. 19, pp. 6891-6897, 1999. http://dx.doi.org/10.1128/mcb.19.10.6891
- P. Sung, and S.A. Stratton, "Yeast Rad51 Recombinase Mediates Polar DNA Strand Exchange in the Absence of ATP Hydrolysis", Journal of Biological Chemistry, vol. 271, pp. 27983-27986, 1996. http://dx.doi.org/10.1074/jbc.271.45.27983
- J.P. Menetski, A. Varghese, and S.C. Kowalczykowski, "Properties of the high-affinity single-stranded DNA binding state of the Escherichia coli recA protein", Biochemistry, vol. 27, pp. 1205-1212, 1988. http://dx.doi.org/10.1021/bi00404a021
- E. Antony, E.J. Tomko, Q. Xiao, L. Krejci, T.M. Lohman, and T. Ellenberger, "Srs2 Disassembles Rad51 Filaments by a Protein-Protein Interaction Triggering ATP Turnover and Dissociation of Rad51 from DNA", Molecular Cell, vol. 35, pp. 105-115, 2009. http://dx.doi.org/10.1016/j.molcel.2009.05.026
- K. Shim, C. Schmutte, K. Yoder, and R. Fishel, "Defining the salt effect on human RAD51 activities", DNA Repair, vol. 5, pp. 718-730, 2006. http://dx.doi.org/10.1016/j.dnarep.2006.03.006
- P. Chi, S. Van Komen, M.G. Sehorn, S. Sigurdsson, and P. Sung, "Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function", DNA Repair, vol. 5, pp. 381-391, 2006. http://dx.doi.org/10.1016/j.dnarep.2005.11.005
- H. Kim, K. Morimatsu, B. Nordén, M. Ardhammar, and M. Takahashi, "ADP stabilizes the human Rad51‐single stranded DNA complex and promotes its DNA annealing activity", Genes to Cells, vol. 7, pp. 1125-1134, 2002. http://dx.doi.org/10.1046/j.1365-2443.2002.00588.x
- S.L. Brenner, R.S. Mitchell, S.W. Morrical, S.K. Neuendorf, B.C. Schutte, and M.M. Cox, "recA protein-promoted ATP hydrolysis occurs throughout recA nucleoprotein filaments.", The Journal of biological chemistry, 1987. http://www.ncbi.nlm.nih.gov/pubmed/2951381
- X. Wang, and J.E. Haber, "Role of Saccharomyces Single-Stranded DNA-Binding Protein RPA in the Strand Invasion Step of Double-Strand Break Repair", PLoS Biology, vol. 2, pp. e21, 2004. http://dx.doi.org/10.1371/journal.pbio.0020021
- T. Sugiyama, and S.C. Kowalczykowski, "Rad52 Protein Associates with Replication Protein A (RPA)-Single-stranded DNA to Accelerate Rad51-mediated Displacement of RPA and Presynaptic Complex Formation", Journal of Biological Chemistry, vol. 277, pp. 31663-31672, 2002. http://dx.doi.org/10.1074/jbc.M203494200
- A. Shinohara, and T. Ogawa, "Stimulation by Rad52 of yeast Rad51- mediated recombination", Nature, vol. 391, pp. 404-407, 1998. http://dx.doi.org/10.1038/34943
- J.H. New, T. Sugiyama, E. Zaitseva, and S.C. Kowalczykowski, "Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A", Nature, vol. 391, pp. 407-410, 1998. http://dx.doi.org/10.1038/34950
- G.T. Milne, and D.T. Weaver, "Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52.", Genes & Development, vol. 7, pp. 1755-1765, 1993. http://dx.doi.org/10.1101/gad.7.9.1755
- A. Shinohara, M. Shinohara, T. Ohta, S. Matsuda, and T. Ogawa, "Rad52 forms ring structures and co‐operates with RPA in single‐strand DNA annealing", Genes to Cells, vol. 3, pp. 145-156, 1998. http://dx.doi.org/10.1046/j.1365-2443.1998.00176.x
- S.L. Hays, A.A. Firmenich, P. Massey, R. Banerjee, and P. Berg, "Studies of the Interaction between Rad52 Protein and the Yeast Single-Stranded DNA Binding Protein RPA", Molecular and Cellular Biology, vol. 18, pp. 4400-4406, 1998. http://dx.doi.org/10.1128/mcb.18.7.4400
- B. Gibb, L.F. Ye, Y. Kwon, H. Niu, P. Sung, and E.C. Greene, "Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules", Nature Structural & Molecular Biology, vol. 21, pp. 893-900, 2014. http://dx.doi.org/10.1038/nsmb.2886
- M.E. Stauffer, and W.J. Chazin, "Physical Interaction between Replication Protein A and Rad51 Promotes Exchange on Single-stranded DNA", Journal of Biological Chemistry, vol. 279, pp. 25638-25645, 2004. http://dx.doi.org/10.1074/jbc.M400029200
- J.H. New, and S.C. Kowalczykowski, "Rad52 Protein Has a Second Stimulatory Role in DNA Strand Exchange That Complements Replication Protein-A Function", Journal of Biological Chemistry, vol. 277, pp. 26171-26176, 2002. http://dx.doi.org/10.1074/jbc.M203670200
- N. Sugawara, X. Wang, and J.E. Haber, "In Vivo Roles of Rad52, Rad54, and Rad55 Proteins in Rad51-Mediated Recombination", Molecular Cell, vol. 12, pp. 209-219, 2003. http://dx.doi.org/10.1016/s1097-2765(03)00269-7
- T. Miyazaki, D.A. Bressan, M. Shinohara, J.E. Haber, and A. Shinohara, "In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair", The EMBO Journal, vol. 23, pp. 939-949, 2004. http://dx.doi.org/10.1038/sj.emboj.7600091
- M. Lisby, J.H. Barlow, R.C. Burgess, and R. Rothstein, "Choreography of the DNA Damage Response", Cell, vol. 118, pp. 699-713, 2004. http://dx.doi.org/10.1016/j.cell.2004.08.015
- T. Rijkers, J. Van Den Ouweland, B. Morolli, A.G. Rolink, W.M. Baarends, P.P.H. Van Sloun, P.H.M. Lohman, and A. Pastink, "Targeted Inactivation of Mouse RAD52Reduces Homologous Recombination but Not Resistance to Ionizing Radiation", Molecular and Cellular Biology, vol. 18, pp. 6423-6429, 1998. http://dx.doi.org/10.1128/mcb.18.11.6423
- Y. Yamaguchi-Iwai, E. Sonoda, J. Buerstedde, O. Bezzubova, C. Morrison, M. Takata, A. Shinohara, and S. Takeda, "Homologous Recombination, but Not DNA Repair, Is Reduced in Vertebrate Cells Deficient in RAD52", Molecular and Cellular Biology, vol. 18, pp. 6430-6435, 1998. http://dx.doi.org/10.1128/mcb.18.11.6430
- Y. Kan, N.N. Batada, and E.A. Hendrickson, "Human somatic cells deficient for RAD52 are impaired for viral integration and compromised for most aspects of homology-directed repair", DNA Repair, vol. 55, pp. 64-75, 2017. http://dx.doi.org/10.1016/j.dnarep.2017.04.006
- A.A. Davies, J. Masson, M.J. McIlwraith, A.Z. Stasiak, A. Stasiak, A.R. Venkitaraman, and S.C. West, "Role of BRCA2 in Control of the RAD51 Recombination and DNA Repair Protein", Molecular Cell, vol. 7, pp. 273-282, 2001. http://dx.doi.org/10.1016/s1097-2765(01)00175-7
- R.B. Jensen, A. Carreira, and S.C. Kowalczykowski, "Purified human BRCA2 stimulates RAD51-mediated recombination", Nature, vol. 467, pp. 678-683, 2010. http://dx.doi.org/10.1038/nature09399
- R. Scully, J. Chen, A. Plug, Y. Xiao, D. Weaver, J. Feunteun, T. Ashley, and D.M. Livingston, "Association of BRCA1 with Rad51 in Mitotic and Meiotic Cells", Cell, vol. 88, pp. 265-275, 1997. http://dx.doi.org/10.1016/s0092-8674(00)81847-4
- J.M.S. Wong, D. Ionescu, and C.J. Ingles, "Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2", Oncogene, vol. 22, pp. 28-33, 2003. http://dx.doi.org/10.1038/sj.onc.1206071
- A. Carreira, J. Hilario, I. Amitani, R.J. Baskin, M.K. Shivji, A.R. Venkitaraman, and S.C. Kowalczykowski, "The BRC Repeats of BRCA2 Modulate the DNA-Binding Selectivity of RAD51", Cell, vol. 136, pp. 1032-1043, 2009. http://dx.doi.org/10.1016/j.cell.2009.02.019
- A.A. Kelso, S.D. Goodson, L.E. Watts, L.L. Ledford, S.M. Waldvogel, J.N. Diehl, S.B. Shah, A.F. Say, J.D. White, and M.G. Sehorn, "The β-isoform of BCCIP promotes ADP release from the RAD51 presynaptic filament and enhances homologous DNA pairing", Nucleic Acids Research, vol. 45, pp. 711-725, 2016. http://dx.doi.org/10.1093/nar/gkw877
- D.V. Bugreev, and A.V. Mazin, "Ca 2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity", Proceedings of the National Academy of Sciences, vol. 101, pp. 9988-9993, 2004. http://dx.doi.org/10.1073/pnas.0402105101
- D.V. Bugreev, F. Huang, O.M. Mazina, R.J. Pezza, O.N. Voloshin, R. Daniel Camerini-Otero, and A.V. Mazin, "HOP2-MND1 modulates RAD51 binding to nucleotides and DNA", Nature Communications, vol. 5, 2014. http://dx.doi.org/10.1038/ncomms5198
- Z. Feng, S.P. Scott, W. Bussen, G.G. Sharma, G. Guo, T.K. Pandita, and S.N. Powell, "Rad52 inactivation is synthetically lethal with BRCA2 deficiency", Proceedings of the National Academy of Sciences, vol. 108, pp. 686-691, 2010. http://dx.doi.org/10.1073/pnas.1010959107
- G.S. Fortin, "Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes", The EMBO Journal, vol. 21, pp. 3160-3170, 2002. http://dx.doi.org/10.1093/emboj/cdf293
- S.L. Hays, A.A. Firmenich, and P. Berg, "Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins.", Proceedings of the National Academy of Sciences, vol. 92, pp. 6925-6929, 1995. http://dx.doi.org/10.1073/pnas.92.15.6925
- X. Veaute, J. Jeusset, C. Soustelle, S.C. Kowalczykowski, E. Le Cam, and F. Fabre, "The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments", Nature, vol. 423, pp. 309-312, 2003. http://dx.doi.org/10.1038/nature01585
- L. Krejci, S. Van Komen, Y. Li, J. Villemain, M.S. Reddy, H. Klein, T. Ellenberger, and P. Sung, "DNA helicase Srs2 disrupts the Rad51 presynaptic filament", Nature, vol. 423, pp. 305-309, 2003. http://dx.doi.org/10.1038/nature01577
- K. Kaniecki, L. De Tullio, B. Gibb, Y. Kwon, P. Sung, and E.C. Greene, "Dissociation of Rad51 Presynaptic Complexes and Heteroduplex DNA Joints by Tandem Assemblies of Srs2", Cell Reports, vol. 21, pp. 3166-3177, 2017. http://dx.doi.org/10.1016/j.celrep.2017.11.047
- C.W. Fung, A.M. Mozlin, and L.S. Symington, "Suppression of the Double-Strand-Break-Repair Defect of the Saccharomyces cerevisiae rad57 Mutant", Genetics, vol. 181, pp. 1195-1206, 2009. http://dx.doi.org/10.1534/genetics.109.100842
- R.P.S. Onge, R. Mani, J. Oh, M. Proctor, E. Fung, R.W. Davis, C. Nislow, F.P. Roth, and G. Giaever, "Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions", Nature Genetics, vol. 39, pp. 199-206, 2007. http://dx.doi.org/10.1038/ng1948
- J. Chun, E.S. Buechelmaier, and S.N. Powell, "Rad51 Paralog Complexes BCDX2 and CX3 Act at Different Stages in the BRCA1-BRCA2-Dependent Homologous Recombination Pathway", Molecular and Cellular Biology, vol. 33, pp. 387-395, 2013. http://dx.doi.org/10.1128/MCB.00465-12
- J. Thacker, "A surfeit of RAD51-like genes?", Trends in Genetics, vol. 15, pp. 166-168, 1999. http://dx.doi.org/10.1016/s0168-9525(99)01733-3
- J.P. Braybrooke, K.G. Spink, J. Thacker, and I.D. Hickson, "The RAD51 Family Member, RAD51L3, Is a DNA-stimulated ATPase That Forms a Complex with XRCC2", Journal of Biological Chemistry, vol. 275, pp. 29100-29106, 2000. http://dx.doi.org/10.1074/jbc.M002075200
- Y. Tsutsui, T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa, "A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene.", Genetics, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10747044
- J. Masson, M.C. Tarsounas, A.Z. Stasiak, A. Stasiak, R. Shah, M.J. McIlwraith, F.E. Benson, and S.C. West, "Identification and purification of two distinct complexes containing the five RAD51 paralogs", Genes & Development, vol. 15, pp. 3296-3307, 2001. http://dx.doi.org/10.1101/gad.947001
- A. Rodrigue, M. Lafrance, M. Gauthier, D. McDonald, M. Hendzel, S.C. West, M. Jasin, and J. Masson, "Interplay between human DNA repair proteins at a unique double-strand break in vivo", The EMBO Journal, vol. 25, pp. 222-231, 2006. http://dx.doi.org/10.1038/sj.emboj.7600914
- Y. Liu, M. Tarsounas, P. O'Regan, and S.C. West, "Role of RAD51C and XRCC3 in Genetic Recombination and DNA Repair", Journal of Biological Chemistry, vol. 282, pp. 1973-1979, 2007. http://dx.doi.org/10.1074/jbc.M609066200
- E. Shor, J. Weinstein, and R. Rothstein, "A Genetic Screen for top3 Suppressors in Saccharomyces cerevisiae Identifies SHU1, SHU2, PSY3 and CSM2", Genetics, vol. 169, pp. 1275-1289, 2005. http://dx.doi.org/10.1534/genetics.104.036764
- H. Sasanuma, M.S. Tawaramoto, J.P. Lao, H. Hosaka, E. Sanda, M. Suzuki, E. Yamashita, N. Hunter, M. Shinohara, A. Nakagawa, and A. Shinohara, "A new protein complex promoting the assembly of Rad51 filaments", Nature Communications, vol. 4, 2013. http://dx.doi.org/10.1038/ncomms2678
- W.A. Gaines, S.K. Godin, F.F. Kabbinavar, T. Rao, A.P. VanDemark, P. Sung, and K.A. Bernstein, "Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52", Nature Communications, vol. 6, 2015. http://dx.doi.org/10.1038/ncomms8834
- K.A. Bernstein, R.J. Reid, I. Sunjevaric, K. Demuth, R.C. Burgess, and R. Rothstein, "The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase", Molecular Biology of the Cell, vol. 22, pp. 1599-1607, 2011. http://dx.doi.org/10.1091/mbc.E10-08-0691
- P. Uetz, L. Giot, G. Cagney, T.A. Mansfield, R.S. Judson, J.R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J.M. Rothberg, "A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae", Nature, vol. 403, pp. 623-627, 2000. http://dx.doi.org/10.1038/35001009
- T. Liu, L. Wan, Y. Wu, J. Chen, and J. Huang, "hSWS1·SWSAP1 Is an Evolutionarily Conserved Complex Required for Efficient Homologous Recombination Repair", Journal of Biological Chemistry, vol. 286, pp. 41758-41766, 2011. http://dx.doi.org/10.1074/jbc.M111.271080
- V. Martín, C. Chahwan, H. Gao, V. Blais, J. Wohlschlegel, J.R. Yates, C.H. McGowan, and P. Russell, "Sws1 is a conserved regulator of homologous recombination in eukaryotic cells", The EMBO Journal, vol. 25, pp. 2564-2574, 2006. http://dx.doi.org/10.1038/sj.emboj.7601141
- A.V. Mazin, A.A. Alexeev, and S.C. Kowalczykowski, "A Novel Function of Rad54 Protein", Journal of Biological Chemistry, vol. 278, pp. 14029-14036, 2003. http://dx.doi.org/10.1074/jbc.M212779200
- B. Wolner, and C.L. Peterson, "ATP-dependent and ATP-independent Roles for the Rad54 Chromatin Remodeling Enzyme during Recombinational Repair of a DNA Double Strand Break", Journal of Biological Chemistry, vol. 280, pp. 10855-10860, 2005. http://dx.doi.org/10.1074/jbc.M414388200
- S. Agarwal, W.A. van Cappellen, A. Guénolé, B. Eppink, S.E. Linsen, E. Meijering, A. Houtsmuller, R. Kanaar, and J. Essers, "ATP-dependent and independent functions of Rad54 in genome maintenance", Journal of Cell Biology, vol. 192, pp. 735-750, 2011. http://dx.doi.org/10.1083/jcb.201011025
- J.A. Solinger, K. Kiianitsa, and W. Heyer, "Rad54, a Swi2/Snf2-like Recombinational Repair Protein, Disassembles Rad51:dsDNA Filaments", Molecular Cell, vol. 10, pp. 1175-1188, 2002. http://dx.doi.org/10.1016/s1097-2765(02)00743-8
- J.M. Mason, K. Dusad, W.D. Wright, J. Grubb, B. Budke, W. Heyer, P.P. Connell, R.R. Weichselbaum, and D.K. Bishop, "RAD54 family translocases counter genotoxic effects of RAD51 in human tumor cells", Nucleic Acids Research, vol. 43, pp. 3180-3196, 2015. http://dx.doi.org/10.1093/nar/gkv175
- J. Spies, A. Waizenegger, O. Barton, M. Sürder, W. Wright, W. Heyer, and M. Löbrich, "Nek1 Regulates Rad54 to Orchestrate Homologous Recombination and Replication Fork Stability", Molecular Cell, vol. 62, pp. 903-917, 2016. http://dx.doi.org/10.1016/j.molcel.2016.04.032
- A. Hayase, M. Takagi, T. Miyazaki, H. Oshiumi, M. Shinohara, and A. Shinohara, "A Protein Complex Containing Mei5 and Sae3 Promotes the Assembly of the Meiosis-Specific RecA Homolog Dmc1", Cell, vol. 119, pp. 927-940, 2004. http://dx.doi.org/10.1016/j.cell.2004.10.031
- F.K. Khasanov, A.F. Salakhova, O.S. Khasanova, A.L. Grishchuk, O.V. Chepurnaja, V.G. Korolev, J. Kohli, and V.I. Bashkirov, "Genetic analysis reveals different roles of Schizosaccharomyces pombe sfr1/dds20 in meiotic and mitotic DNA recombination and repair", Current Genetics, vol. 54, pp. 197-211, 2008. http://dx.doi.org/10.1007/s00294-008-0212-z
- J. Yuan, and J. Chen, "The Role of the Human SWI5-MEI5 Complex in Homologous Recombination Repair", Journal of Biological Chemistry, vol. 286, pp. 9888-9893, 2011. http://dx.doi.org/10.1074/jbc.M110.207290
- S. Tsai, G. Su, S. Lin, C. Chung, X. Xue, M.H. Dunlop, Y. Akamatsu, M. Jasin, P. Sung, and P. Chi, "Rad51 presynaptic filament stabilization function of the mouse Swi5–Sfr1 heterodimeric complex", Nucleic Acids Research, vol. 40, pp. 6558-6569, 2012. http://dx.doi.org/10.1093/nar/gks305
- Y. Kokabu, Y. Murayama, N. Kuwabara, T. Oroguchi, H. Hashimoto, Y. Tsutsui, N. Nozaki, S. Akashi, S. Unzai, T. Shimizu, H. Iwasaki, M. Sato, and M. Ikeguchi, "Fission Yeast Swi5-Sfr1 Protein Complex, an Activator of Rad51 Recombinase, Forms an Extremely Elongated Dogleg-shaped Structure", Journal of Biological Chemistry, vol. 286, pp. 43569-43576, 2011. http://dx.doi.org/10.1074/jbc.M111.303339
- N. Haruta, Y. Kurokawa, Y. Murayama, Y. Akamatsu, S. Unzai, Y. Tsutsui, and H. Iwasaki, "The Swi5–Sfr1 complex stimulates Rhp51/Rad51 - and Dmc1-mediated DNA strand exchange in vitro", Nature Structural & Molecular Biology, vol. 13, pp. 823-830, 2006. http://dx.doi.org/10.1038/nsmb1136
-
H.E. Alatwi, and J.A. Downs, "Removal of H2A.Z by
INO 80 promotes homologous recombination", EMBO reports, vol. 16, pp. 986-994, 2015. http://dx.doi.org/10.15252/embr.201540330 - C.A. Lademann, J. Renkawitz, B. Pfander, and S. Jentsch, "The INO80 Complex Removes H2A.Z to Promote Presynaptic Filament Formation during Homologous Recombination", Cell Reports, vol. 19, pp. 1294-1303, 2017. http://dx.doi.org/10.1016/j.celrep.2017.04.051
- T. Tsukuda, A.B. Fleming, J.A. Nickoloff, and M.A. Osley, "Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae", Nature, vol. 438, pp. 379-383, 2005. http://dx.doi.org/10.1038/nature04148
- L. Chen, Y. Cai, J. Jin, L. Florens, S.K. Swanson, M.P. Washburn, J.W. Conaway, and R.C. Conaway, "Subunit Organization of the Human INO80 Chromatin Remodeling Complex", Journal of Biological Chemistry, vol. 286, pp. 11283-11289, 2011. http://dx.doi.org/10.1074/jbc.M111.222505
- M. Papamichos-Chronakis, J.E. Krebs, and C.L. Peterson, "Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptationin response to DNA damage", Genes & Development, vol. 20, pp. 2437-2449, 2006. http://dx.doi.org/10.1101/gad.1440206
- H. van Attikum, O. Fritsch, and S.M. Gasser, "Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks", The EMBO Journal, vol. 26, pp. 4113-4125, 2007. http://dx.doi.org/10.1038/sj.emboj.7601835
- P.P. Shah, X. Zheng, A. Epshtein, J.N. Carey, D.K. Bishop, and H.L. Klein, "Swi2/Snf2-Related Translocases Prevent Accumulation of Toxic Rad51 Complexes during Mitotic Growth", Molecular Cell, vol. 39, pp. 862-872, 2010. http://dx.doi.org/10.1016/j.molcel.2010.08.028
- P. Chi, Y. Kwon, C. Seong, A. Epshtein, I. Lam, P. Sung, and H.L. Klein, "Yeast Recombination Factor Rdh54 Functionally Interacts with the Rad51 Recombinase and Catalyzes Rad51 Removal from DNA", Journal of Biological Chemistry, vol. 281, pp. 26268-26279, 2006. http://dx.doi.org/10.1074/jbc.M602983200
- M. Raschle, S. Van Komen, P. Chi, T. Ellenberger, and P. Sung, "Multiple Interactions with the Rad51 Recombinase Govern the Homologous Recombination Function of Rad54", Journal of Biological Chemistry, vol. 279, pp. 51973-51980, 2004. http://dx.doi.org/10.1074/jbc.M410101200
- S.R. Santa Maria, Y. Kwon, P. Sung, and H.L. Klein, "Characterization of the Interaction between the Saccharomyces cerevisiae Rad51 Recombinase and the DNA Translocase Rdh54", Journal of Biological Chemistry, vol. 288, pp. 21999-22005, 2013. http://dx.doi.org/10.1074/jbc.M113.480475
- P. Chi, Y. Kwon, M. Visnapuu, I. Lam, S.R. Santa Maria, X. Zheng, A. Epshtein, E.C. Greene, P. Sung, and H.L. Klein, "Analyses of the yeast Rad51 recombinase A265V mutant reveal different in vivo roles of Swi2-like factors", Nucleic Acids Research, vol. 39, pp. 6511-6522, 2011. http://dx.doi.org/10.1093/nar/gkr297
- A.V. Mazin, O.M. Mazina, D.V. Bugreev, and M.J. Rossi, "Rad54, the motor of homologous recombination", DNA Repair, vol. 9, pp. 286-302, 2010. http://dx.doi.org/10.1016/j.dnarep.2009.12.006
- E. Boehm, M. Gildenberg, and M. Washington, "The Many Roles of PCNA in Eukaryotic DNA Replication", The Enzymes, pp. 231-254, 2016. http://dx.doi.org/10.1016/bs.enz.2016.03.003
- M.L. Bochman, N. Sabouri, and V.A. Zakian, "Unwinding the functions of the Pif1 family helicases", DNA Repair, vol. 9, pp. 237-249, 2010. http://dx.doi.org/10.1016/j.dnarep.2010.01.008
- B. Pfander, G. Moldovan, M. Sacher, C. Hoege, and S. Jentsch, "SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase", Nature, vol. 436, pp. 428-433, 2005. http://dx.doi.org/10.1038/nature03665
- R.C. Burgess, M. Lisby, V. Altmannova, L. Krejci, P. Sung, and R. Rothstein, "Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo", Journal of Cell Biology, vol. 185, pp. 969-981, 2009. http://dx.doi.org/10.1083/jcb.200810055
- A.A. Armstrong, F. Mohideen, and C.D. Lima, "Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2", Nature, vol. 483, pp. 59-63, 2012. http://dx.doi.org/10.1038/nature10883
- A. Aguilera, and H.L. Klein, "Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations.", Genetics, 1988. http://www.ncbi.nlm.nih.gov/pubmed/3044923
- S. Colavito, M. Macris-Kiss, C. Seong, O. Gleeson, E.C. Greene, H.L. Klein, L. Krejci, and P. Sung, "Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption", Nucleic Acids Research, vol. 37, pp. 6754-6764, 2009. http://dx.doi.org/10.1093/nar/gkp748
- D. Branzei, F. Vanoli, and M. Foiani, "SUMOylation regulates Rad18-mediated template switch", Nature, vol. 456, pp. 915-920, 2008. http://dx.doi.org/10.1038/nature07587
- M. Urulangodi, M. Sebesta, D. Menolfi, B. Szakal, J. Sollier, A. Sisakova, L. Krejci, and D. Branzei, "Local regulation of the Srs2 helicase by the SUMO-like domain protein Esc2 promotes recombination at sites of stalled replication", Genes & Development, vol. 29, pp. 2067-2080, 2015. http://dx.doi.org/10.1101/gad.265629.115
- W. Feng, and M. Jasin, "BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination", Nature Communications, vol. 8, 2017. http://dx.doi.org/10.1038/s41467-017-00634-0
- R. Zellweger, D. Dalcher, K. Mutreja, M. Berti, J.A. Schmid, R. Herrador, A. Vindigni, and M. Lopes, "Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells", Journal of Cell Biology, vol. 208, pp. 563-579, 2015. http://dx.doi.org/10.1083/jcb.201406099
- Y. Hashimoto, A. Ray Chaudhuri, M. Lopes, and V. Costanzo, "Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis", Nature Structural & Molecular Biology, vol. 17, pp. 1305-1311, 2010. http://dx.doi.org/10.1038/nsmb.1927
- A.M. Kolinjivadi, V. Sannino, A. De Antoni, K. Zadorozhny, M. Kilkenny, H. Técher, G. Baldi, R. Shen, A. Ciccia, L. Pellegrini, L. Krejci, and V. Costanzo, "Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments", Molecular Cell, vol. 67, pp. 867-881.e7, 2017. http://dx.doi.org/10.1016/j.molcel.2017.07.001
- K. Schlacher, N. Christ, N. Siaud, A. Egashira, H. Wu, and M. Jasin, "Double-Strand Break Repair-Independent Role for BRCA2 in Blocking Stalled Replication Fork Degradation by MRE11", Cell, vol. 145, pp. 529-542, 2011. http://dx.doi.org/10.1016/j.cell.2011.03.041
- H. Dungrawala, K.P. Bhat, R. Le Meur, W.J. Chazin, X. Ding, S.K. Sharan, S.R. Wessel, A.A. Sathe, R. Zhao, and D. Cortez, "RADX Promotes Genome Stability and Modulates Chemosensitivity by Regulating RAD51 at Replication Forks", Molecular Cell, vol. 67, pp. 374-386.e5, 2017. http://dx.doi.org/10.1016/j.molcel.2017.06.023
- R. González-Prieto, A.M. Muñoz-Cabello, M.J. Cabello-Lobato, and F. Prado, "Rad51 replication fork recruitment is required for DNA damage tolerance", The EMBO Journal, vol. 32, pp. 1307-1321, 2013. http://dx.doi.org/10.1038/emboj.2013.73
- T. Tsurimoto, and B. Stillman, "Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4.", Proceedings of the National Academy of Sciences, vol. 87, pp. 1023-1027, 1990. http://dx.doi.org/10.1073/pnas.87.3.1023
- J. Cai, F. Uhlmann, E. Gibbs, H. Flores-Rozas, C. Lee, B. Phillips, J. Finkelstein, N. Yao, M. O’Donnell, and J. Hurwitz, "Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells", Proceedings of the National Academy of Sciences, vol. 93, pp. 12896-12901, 1996. http://dx.doi.org/10.1073/pnas.93.23.12896
- M.M. Hingorani, and M.M. Coman, "On the Specificity of Interaction between the Saccharomyces cerevisiae Clamp Loader Replication Factor C and Primed DNA Templates during DNA Replication", Journal of Biological Chemistry, vol. 277, pp. 47213-47224, 2002. http://dx.doi.org/10.1074/jbc.M206764200
- T. Tsurimoto, and B. Stillman, "Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis.", The Journal of biological chemistry, 1991. http://www.ncbi.nlm.nih.gov/pubmed/1671046
- X.V. Gomes, and P.M. Burgers, "ATP Utilization by Yeast Replication Factor C", Journal of Biological Chemistry, vol. 276, pp. 34768-34775, 2001. http://dx.doi.org/10.1074/jbc.M011631200
- S. Waga, and B. Stillman, "Cyclin-Dependent Kinase Inhibitor p21 Modulates the DNA Primer-Template Recognition Complex", Molecular and Cellular Biology, vol. 18, pp. 4177-4187, 1998. http://dx.doi.org/10.1128/mcb.18.7.4177
- J. Li, D.L. Holzschu, and T. Sugiyama, "PCNA is efficiently loaded on the DNA recombination intermediate to modulate polymerase δ, η, and ζ activities", Proceedings of the National Academy of Sciences, vol. 110, pp. 7672-7677, 2013. http://dx.doi.org/10.1073/pnas.1222241110
- A. Yuzhakov, "Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme", The EMBO Journal, vol. 18, pp. 6189-6199, 1999. http://dx.doi.org/10.1093/emboj/18.21.6189
- L. De Tullio, K. Kaniecki, Y. Kwon, J.B. Crickard, P. Sung, and E.C. Greene, "Yeast Srs2 Helicase Promotes Redistribution of Single-Stranded DNA-Bound RPA and Rad52 in Homologous Recombination Regulation", Cell Reports, vol. 21, pp. 570-577, 2017. http://dx.doi.org/10.1016/j.celrep.2017.09.073
- G. Ira, A. Malkova, G. Liberi, M. Foiani, and J.E. Haber, "Srs2 and Sgs1–Top3 Suppress Crossovers during Double-Strand Break Repair in Yeast", Cell, vol. 115, pp. 401-411, 2003. http://dx.doi.org/10.1016/s0092-8674(03)00886-9
- T. Miura, T. Shibata, and K. Kusano, "Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity", Proceedings of the National Academy of Sciences, vol. 110, pp. 16067-16072, 2013. http://dx.doi.org/10.1073/pnas.1303111110
- J. Liu, C. Ede, W.D. Wright, S.K. Gore, S.S. Jenkins, B.D. Freudenthal, M. Todd Washington, X. Veaute, and W. Heyer, "Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase δ-extending D-loops", eLife, vol. 6, 2017. http://dx.doi.org/10.7554/eLife.22195
- M. Yeung, and D. Durocher, "Srs2 enables checkpoint recovery by promoting disassembly of DNA damage foci from chromatin", DNA Repair, vol. 10, pp. 1213-1222, 2011. http://dx.doi.org/10.1016/j.dnarep.2011.09.005
- M.B. Vaze, A. Pellicioli, S.E. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J.E. Haber, "Recovery from Checkpoint-Mediated Arrest after Repair of a Double-Strand Break Requires Srs2 Helicase", Molecular Cell, vol. 10, pp. 373-385, 2002. http://dx.doi.org/10.1016/s1097-2765(02)00593-2
- T. van der Heijden, R. Seidel, M. Modesti, R. Kanaar, C. Wyman, and C. Dekker, "Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules", Nucleic Acids Research, vol. 35, pp. 5646-5657, 2007. http://dx.doi.org/10.1093/nar/gkm629
- X. Li, and W. Heyer, "RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae", Nucleic Acids Research, vol. 37, pp. 638-646, 2008. http://dx.doi.org/10.1093/nar/gkn980
- S. Inano, K. Sato, Y. Katsuki, W. Kobayashi, H. Tanaka, K. Nakajima, S. Nakada, H. Miyoshi, K. Knies, A. Takaori-Kondo, D. Schindler, M. Ishiai, H. Kurumizaka, and M. Takata, "RFWD3-Mediated Ubiquitination Promotes Timely Removal of Both RPA and RAD51 from DNA Damage Sites to Facilitate Homologous Recombination", Molecular Cell, vol. 66, pp. 622-634.e8, 2017. http://dx.doi.org/10.1016/j.molcel.2017.04.022
- D.V. Bugreev, X. Yu, E.H. Egelman, and A.V. Mazin, "Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase", Genes & Development, vol. 21, pp. 3085-3094, 2007. http://dx.doi.org/10.1101/gad.1609007
- Y. Hu, S. Raynard, M.G. Sehorn, X. Lu, W. Bussen, L. Zheng, J.M. Stark, E.L. Barnes, P. Chi, P. Janscak, M. Jasin, H. Vogel, P. Sung, and G. Luo, "RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments", Genes & Development, vol. 21, pp. 3073-3084, 2007. http://dx.doi.org/10.1101/gad.1609107
- J. Simandlova, J. Zagelbaum, M.J. Payne, W.K. Chu, I. Shevelev, K. Hanada, S. Chatterjee, D.A. Reid, Y. Liu, P. Janscak, E. Rothenberg, and I.D. Hickson, "FBH1 Helicase Disrupts RAD51 Filaments in Vitro and Modulates Homologous Recombination in Mammalian Cells", Journal of Biological Chemistry, vol. 288, pp. 34168-34180, 2013. http://dx.doi.org/10.1074/jbc.M113.484493
- J.A. Sommers, N. Rawtani, R. Gupta, D.V. Bugreev, A.V. Mazin, S.B. Cantor, and R.M. Brosh, "FANCJ Uses Its Motor ATPase to Destabilize Protein-DNA Complexes, Unwind Triplexes, and Inhibit RAD51 Strand Exchange", Journal of Biological Chemistry, vol. 284, pp. 7505-7517, 2009. http://dx.doi.org/10.1074/jbc.M809019200
- G. Moldovan, D. Dejsuphong, M. Petalcorin, K. Hofmann, S. Takeda, S. Boulton, and A. D'Andrea, "Inhibition of Homologous Recombination by the PCNA-Interacting Protein PARI", Molecular Cell, vol. 45, pp. 75-86, 2012. http://dx.doi.org/10.1016/j.molcel.2011.11.010
- K. Fugger, M. Mistrik, J.R. Danielsen, C. Dinant, J. Falck, J. Bartek, J. Lukas, and N. Mailand, "Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities", Journal of Cell Biology, vol. 186, pp. 655-663, 2009. http://dx.doi.org/10.1083/jcb.200812138
- G. Traverso, C. Bettegowda, J. Kraus, M.R. Speicher, K.W. Kinzler, B. Vogelstein, and C. Lengauer, "Hyper-recombination and genetic instability in BLM-deficient epithelial cells.", Cancer research, 2003. http://www.ncbi.nlm.nih.gov/pubmed/14695165
- Y. Hu, X. Lu, E. Barnes, M. Yan, H. Lou, and G. Luo, "Recql5 and Blm RecQ DNA Helicases Have Nonredundant Roles in Suppressing Crossovers", Molecular and Cellular Biology, vol. 25, pp. 3431-3442, 2005. http://dx.doi.org/10.1128/MCB.25.9.3431-3442.2005
- J.R. Lydeard, Z. Lipkin-Moore, S. Jain, V.V. Eapen, and J.E. Haber, "Sgs1 and Exo1 Redundantly Inhibit Break-Induced Replication and De Novo Telomere Addition at Broken Chromosome Ends", PLoS Genetics, vol. 6, pp. e1000973, 2010. http://dx.doi.org/10.1371/journal.pgen.1000973
- W. Chung, Z. Zhu, A. Papusha, A. Malkova, and G. Ira, "Defective Resection at DNA Double-Strand Breaks Leads to De Novo Telomere Formation and Enhances Gene Targeting", PLoS Genetics, vol. 6, pp. e1000948, 2010. http://dx.doi.org/10.1371/journal.pgen.1000948
- J.H. Barlow, and R. Rothstein, "Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase", The EMBO Journal, vol. 28, pp. 1121-1130, 2009. http://dx.doi.org/10.1038/emboj.2009.43
- M. Lisby, R. Rothstein, and U.H. Mortensen, "Rad52 forms DNA repair and recombination centers during S phase", Proceedings of the National Academy of Sciences, vol. 98, pp. 8276-8282, 2001. http://dx.doi.org/10.1073/pnas.121006298
- L.J. Holt, B.B. Tuch, J. Villén, A.D. Johnson, S.P. Gygi, and D.O. Morgan, "Global Analysis of Cdk1 Substrate Phosphorylation Sites Provides Insights into Evolution", Science, vol. 325, pp. 1682-1686, 2009. http://dx.doi.org/10.1126/science.1172867
- M. Sacher, B. Pfander, C. Hoege, and S. Jentsch, "Control of Rad52 recombination activity by double-strand break-induced SUMO modification", Nature Cell Biology, vol. 8, pp. 1284-1290, 2006. http://dx.doi.org/10.1038/ncb1488
- V. Altmannova, N. Eckert-Boulet, M. Arneric, P. Kolesar, R. Chaloupkova, J. Damborsky, P. Sung, X. Zhao, M. Lisby, and L. Krejci, "Rad52 SUMOylation affects the efficiency of the DNA repair", Nucleic Acids Research, vol. 38, pp. 4708-4721, 2010. http://dx.doi.org/10.1093/nar/gkq195
- V.I. Bashkirov, J.S. King, E.V. Bashkirova, J. Schmuckli-Maurer, and W. Heyer, "DNA Repair Protein Rad55 Is a Terminal Substrate of the DNA Damage Checkpoints", Molecular and Cellular Biology, vol. 20, pp. 4393-4404, 2000. http://dx.doi.org/10.1128/mcb.20.12.4393-4404.2000
- K. Herzberg, V.I. Bashkirov, M. Rolfsmeier, E. Haghnazari, W.H. McDonald, S. Anderson, E.V. Bashkirova, J.R. Yates, and W. Heyer, "Phosphorylation of Rad55 on Serines 2, 8, and 14 Is Required for Efficient Homologous Recombination in the Recovery of Stalled Replication Forks", Molecular and Cellular Biology, vol. 26, pp. 8396-8409, 2006. http://dx.doi.org/10.1128/MCB.01317-06
- S. Flott, Y. Kwon, Y.Z. Pigli, P.A. Rice, P. Sung, and S.P. Jackson, "Regulation of Rad51 function by phosphorylation", EMBO reports, vol. 12, pp. 833-839, 2011. http://dx.doi.org/10.1038/embor.2011.127
- H. Dou, C. Huang, M. Singh, P.B. Carpenter, and E.T. Yeh, "Regulation of DNA Repair through DeSUMOylation and SUMOylation of Replication Protein A Complex", Molecular Cell, vol. 39, pp. 333-345, 2010. http://dx.doi.org/10.1016/j.molcel.2010.07.021
- C. Cremona, P. Sarangi, Y. Yang, L. Hang, S. Rahman, and X. Zhao, "Extensive DNA Damage-Induced Sumoylation Contributes to Replication and Repair and Acts in Addition to the Mec1 Checkpoint", Molecular Cell, vol. 45, pp. 422-432, 2012. http://dx.doi.org/10.1016/j.molcel.2011.11.028
- I. Psakhye, and S. Jentsch, "Protein Group Modification and Synergy in the SUMO Pathway as Exemplified in DNA Repair", Cell, vol. 151, pp. 807-820, 2012. http://dx.doi.org/10.1016/j.cell.2012.10.021
- H. Shima, H. Suzuki, J. Sun, K. Kono, L. Shi, A. Kinomura, Y. Horikoshi, T. Ikura, M. Ikura, R. Kanaar, K. Igarashi, H. Saitoh, H. Kurumizaka, and S. Tashiro, "Activation of the SUMO modification system is required for the accumulation of RAD51 at sites containing DNA damage", Journal of Cell Science, 2013. http://dx.doi.org/10.1242/jcs.133744
- K. Luo, L. Li, Y. Li, C. Wu, Y. Yin, Y. Chen, M. Deng, S. Nowsheen, J. Yuan, and Z. Lou, "A phosphorylation–deubiquitination cascade regulates the BRCA2–RAD51 axis in homologous recombination", Genes & Development, vol. 30, pp. 2581-2595, 2016. http://dx.doi.org/10.1101/gad.289439.116
- J. Liu, A. Kruswick, H. Dang, A.D. Tran, S.M. Kwon, X.W. Wang, and P. Oberdoerffer, "Ubiquitin-specific protease 21 stabilizes BRCA2 to control DNA repair and tumor growth", Nature Communications, vol. 8, 2017. http://dx.doi.org/10.1038/s41467-017-00206-2
- I. Chiolo, M. Saponaro, A. Baryshnikova, J. Kim, Y. Seo, and G. Liberi, "The Human F-Box DNA Helicase FBH1 Faces Saccharomyces cerevisiae Srs2 and Postreplication Repair Pathway Roles", Molecular and Cellular Biology, vol. 27, pp. 7439-7450, 2007. http://dx.doi.org/10.1128/MCB.00963-07
- M. Saponaro, D. Callahan, X. Zheng, L. Krejci, J.E. Haber, H.L. Klein, and G. Liberi, "Cdk1 Targets Srs2 to Complete Synthesis-Dependent Strand Annealing and to Promote Recombinational Repair", PLoS Genetics, vol. 6, pp. e1000858, 2010. http://dx.doi.org/10.1371/journal.pgen.1000858
- P. Kolesar, P. Sarangi, V. Altmannova, X. Zhao, and L. Krejci, "Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation", Nucleic Acids Research, vol. 40, pp. 7831-7843, 2012. http://dx.doi.org/10.1093/nar/gks484
ACKNOWLEDGMENTS
We are grateful to Benura Azeroglu, Elizabeth Bayne and Sara Buonomo for critical reading of the manuscript and helpful suggestions. T.A. thanks BBSRC and the School of Biological Sciences, University of Edinburgh for an EastBio doctoral training partnership stipend (grant number BB/M010996/1). O.K. is a recipient of an Edinburgh Global Research Scholarship from the School of Biological Sciences. The Makovets lab is funded by the Leverhulme Trust Project Grant RPG-2016-379 to S.M.
COPYRIGHT
© 2018

Putting together and taking apart: assembly and disassembly of the Rad51 nucleoprotein filament in DNA repair and genome stability by Andriuskevicius et al. is licensed under a Creative Commons Attribution 4.0 International License.