Back to article: A game of substrates: replication fork remodeling and its roles in genome stability and chemo-resistance
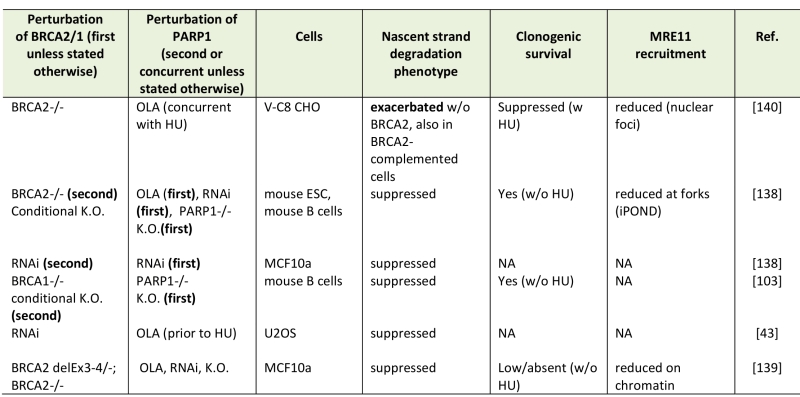
TABLE 2. Nascent strand degradation phenotypes of BRCA-deficient, PARP1-deficient cells.
Abbreviations: OLA, olaparib, K.O., knockout, RNAi, siRNA or shRNA-mediated depletion, HU, hydroxyurea.
43. Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Ray Chaudhuri A, Nussenzweig A, Janscak P, Lopes M (2017). Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun 8(1): 859. http://dx.doi.org/10.1038/s41467-017-01164-5
103. Chaudhuri RA, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, John S, Day A, Crespo AV, Shen B, Starnes LM, de Ruiter JR, Daniel JA, Konstantinopoulos PA, Cortez D, Cantor SB, Fernandez-Capetillo O, Ge K, Jonkers J, Rottenberg S, Sharan SK, Nussenzweig A (2016). Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535(7612): 382-387. http://dx.doi.org/10.1038/nature18325
138. Ding X, Ray Chaudhuri A, Callen E, Pang Y, Biswas K, Klarmann KD, Martin BK, Burkett S, Cleveland L, Stauffer S, Sullivan T, Dewan A, Marks H, Tubbs AT, Wong N, Buehler E, Akagi K, Martin SE, Keller JR, Nussenzweig A, Sharan SK (2016). Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat Commun 7:12425. http://dx.doi.org/10.1038/ncomms12425
139. Feng W, Jasin M (2017). BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat Commun 8(1): 525. http://dx.doi.org/10.1038/s41467-017-00634-0
140. Ying S, Chen Z, Medhurst AL, Neal JA, Bao Z, Mortusewicz O, McGouran J, Song X, Shen H, Hamdy FC, Kessler BM, Meek K, Helleday T (2016). DNA-PKcs and PARP1 Bind to Unresected Stalled DNA Replication Forks Where They Recruit XRCC1 to Mediate Repair. Cancer Res 76(5): 1078-1088. http://dx.doi.org/10.1158/0008-5472.can-15-0608