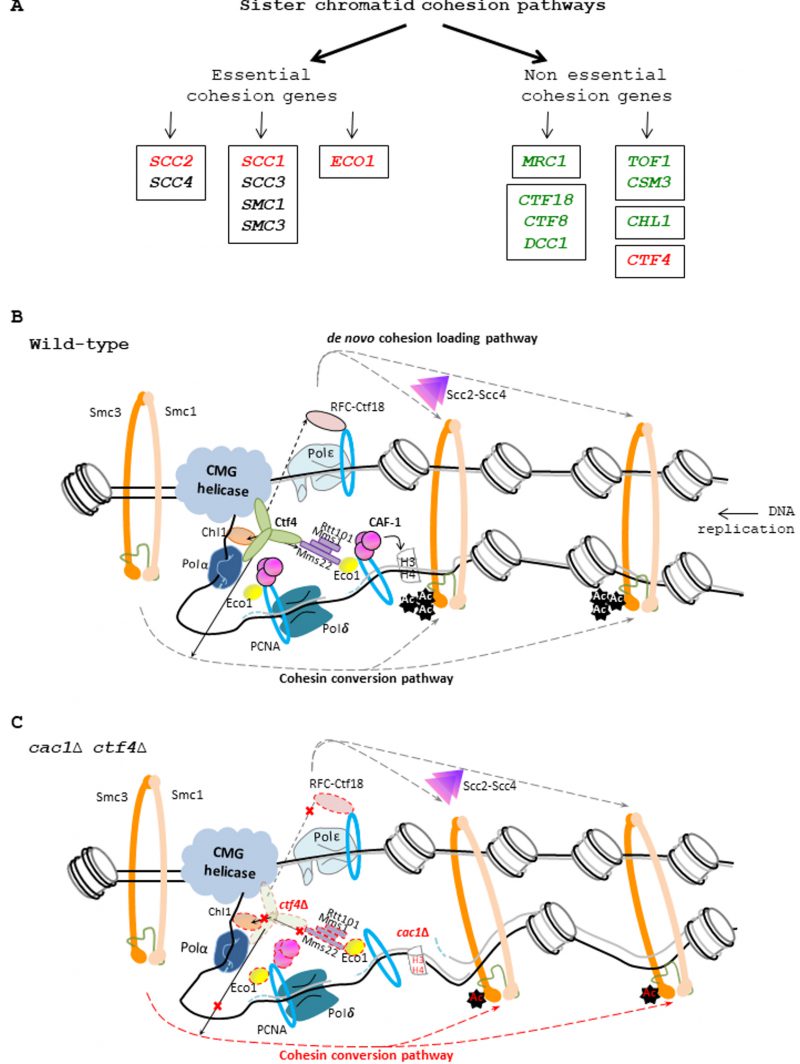
FIGURE 9: CAF-1 and the replisomal protein Ctf4 are important for cohesion. (A) Genetic interactions between CAF-1 complex and genes involved in sister chromatid cohesion. Left, essential cohesion genes. Right, non-essential cohesion genes encoding replisome-associated proteins that are essential for efficient Smc3 acetylation by Eco1, and for cohesion establishment [69] [121]. The Mrc1, CTF18-RFC pathway is involved in de novo loading of cohesins onto nascent DNAs [74]. The Tof1/Csm3, Chl1, Ctf4 pathway is involved in conversion of chromosome associated cohesins into cohesive structure during S phase [74]. Genes enclosed encode protein to form a complex. Genes in green are not required for growth in absence of CAF-1 function. Genes in red are required for growth in absence of CAF-1 function. For essential genes, we used thermosensitive mutants to analyze the genetic interactions with CAF-1 mutants. Genes in black: Genetic interaction with CAF-1 mutants non-determined. (B) Illustration of the role played by Ctf4 in the SCC establishment. Ctf4 links DNA replication with sister chromatid cohesion establishment by recruiting Chl1 helicase to the replisome where it directly interacts with cohesins and assists Eco1, which acetylates Smc3, in replication coupled-cohesin establishment [38] [77]. Ctf4 interacts and tethers the Rtt101-Mms1-Mms22 E3 ubiquitin ligase to the replisome which in turn recruits and/or promotes Eco1-dependent sister chromatid cohesion [37] [86]. Ctf4 can also help the chromatin recruitment of Ctf18-RFC [56] a complex that, loads and unloads PCNA, could act as a binding platform for recruiting Eco1, and is involved with the cohesin loader Scc2-Scc4 in the de novo loading of cohesins onto nascent DNA [74]. Finally, Ctf4 is also required for the conversion of cohesins rings preloaded onto the DNA template into a cohesive form [74]. (C) Proposed model explaining the importance of CAF-1 in ctf4Δ cells. In absence of Ctf4, multiple cohesion establishment pathways are affected (red cross) leading to major defects in SCC establishment. In absence of CAF-1 fewer nucleosomes are deposited on replicated DNA, generating fewer but longer Okazaki fragments [140], leading to increased inter-nucleosome spacing in nascent chromatin and inappropriate epigenetic states [13] [148]. In these conditions, the nucleosome assembly function of CAF-1 is required to maintain SCC and efficient cell growth in yeast affected in cohesion. In ctf4Δ cac1Δ cells, the altered chromatin structure arising in absence of CAF-1 function increases the SCC defects induced by the absence of Ctf4, leading to severe growth defects and genomic instability.