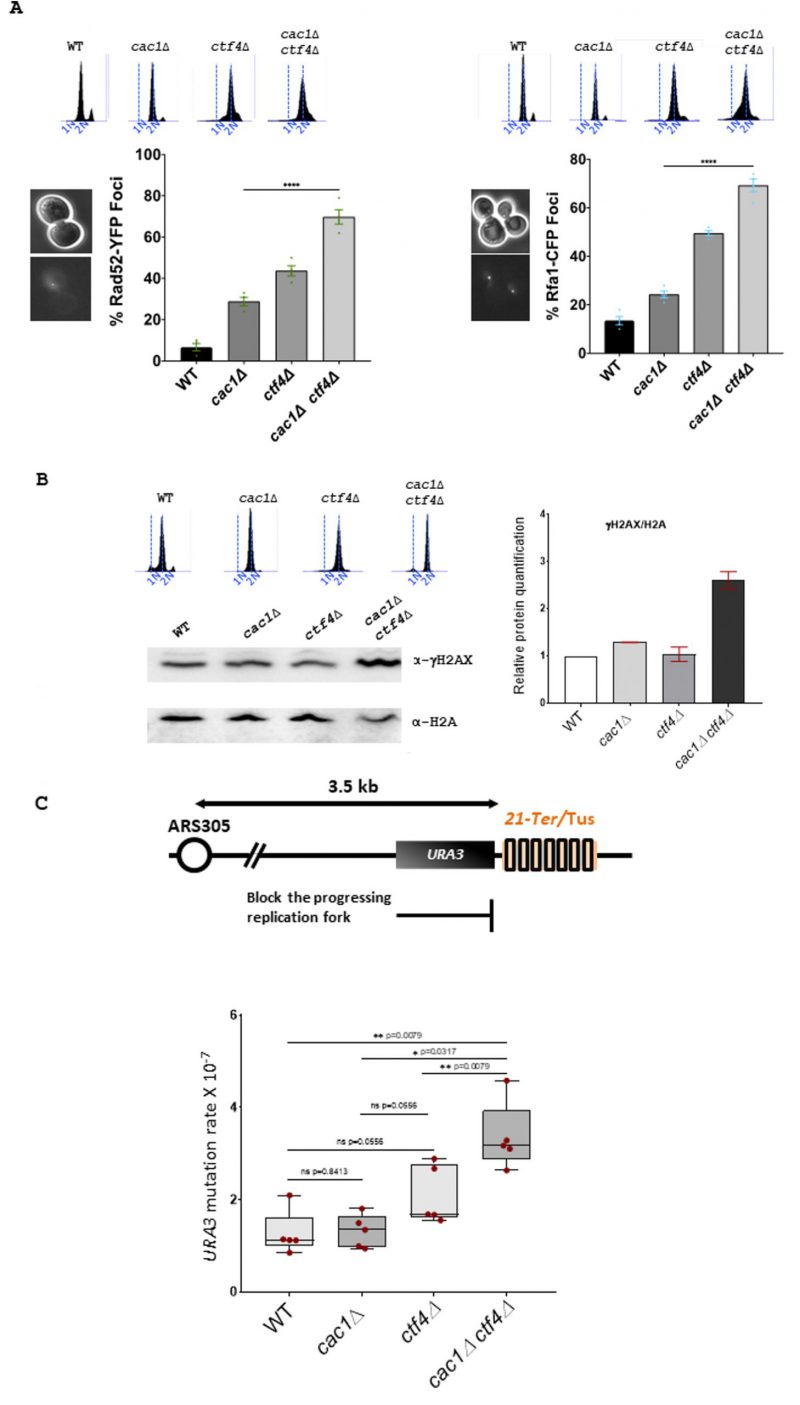
FIGURE 3: Genomic integrity is affected in cac1Δ ctf4Δ cells. (A) Left, Rad52 foci are increased in cac1Δ ctf4Δ cells. Wild-type, cac1Δ, ctf4Δ, and cac1Δ ctf4Δ cells encoding Rad52-YFP were analyzed with fluorescence microscopy. Right, Rfa1 foci are increased in cac1Δ ctf4Δ cells. Wild-type, cac1Δ, ctf4Δ, and cac1Δ ctf4Δ cells encoding Rfa1-CFP were analyzed with fluorescence microscopy. Numbers indicate the percentage of cells that contained Rad52-YFP (left) or Rfa1-CFP foci (right). DNA replication was monitored by FACS analysis of DNA content. At least 200 cells were analyzed for each strain from three independent experiments. Statistical significance was measured using the two-tailed Mann-Whitney test. (B) γH2AX is increased in the absence of Cac1 in ctf4Δ cells. Left, Western blot was used to detect phosphorylation of H2A serine 129 (γH2AX). Right, histogram shows for WT, cac1Δ, ctf4Δ, and cac1Δ ctf4Δ, γH2AX/H2A ratios calculated based on Western blots signal intensities. DNA replication was monitored by FACS analysis of DNA content. The experiment has been done in duplicate. (C) CAF-1 inactivation increases mutation rate in ctf4Δ cells at the Tus/Ter barrier. Top, Schematic representation of the unidirectional and site-specific Tus-Ter replication fork barrier. Ter sequence is integrated 3.5 kb downstream ARS305 on Chromosome III where Tus protein (colored rectangles) binds specifically to Ter sequence, causing replication fork pausing. Upstream to Tus/Ter replication fork barrier is the URA3 reporter gene, which permits the positive selection for ura3 mutations in presence of 5-FOA to measure mutation rate. Bottom, exponentially growing cells expressing Tus protein were plated for 3 days at 30° on YPGal plates and plated out on 5-FOA to select for ura3 mutation. Box-and-whisker plots, representing the upper and lower quartile with the median, show the mutation rate in WT, cac1Δ, ctf4Δ, and cac1Δ ctf4Δ cells. Statistical analyses were done on n=5 independent experiments using two-tailed Mann-Whitney test; *p<0.05; ** p< 0.005; **** p<0.0001; ns, not significant.