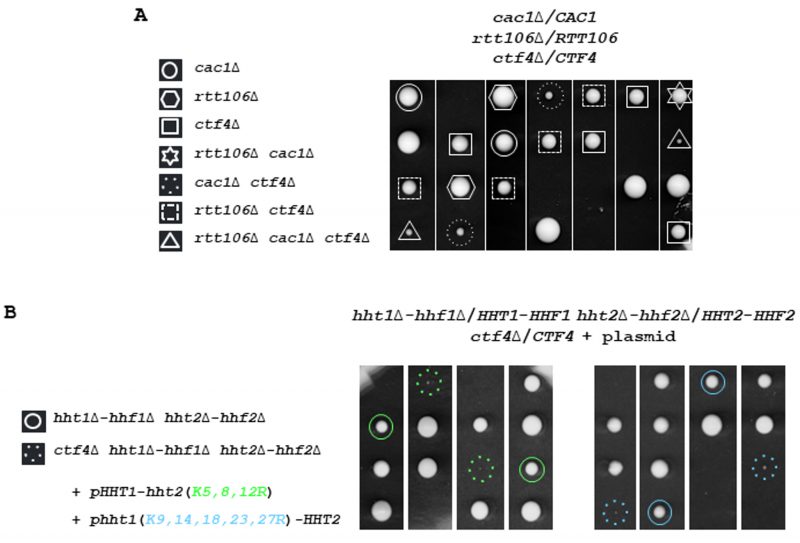
FIGURE 1: CTF4 inactivation results in cell lethality in different genetic contexts affecting chromatin assembly. (A) Defective Cac1/Rtt106-dependent chromatin assembly affects growth in absence of CTF4. Tetrads from rtt106Δ/RTT106 cac1Δ/CAC1 ctf4Δ/CTF4 diploid strain were dissected. In this and subsequent figures, the spores from a given tetrad are in vertical line in a YPD plate. Fifty tetrads were dissected. Five representative tetrads are shown after 3 days at 30°. (B) Mutations at histone lysine residues implicated in nucleosome assembly strongly affect growth of ctf4Δ cells. One hundred tetrads from diploids for hht1Δ-hhf1Δ/HHT1-HHF1 hht2Δ-hhf2Δ/HHT2-HHF2 ctf4Δ/CTF4 expressing either HHT2 and hhf2(K5,8,12R), or HHF1 and hht1(K9,14,18,23,27R) from a centromeric plasmid were dissected and analyzed for the presence of auxotrophic markers. The circle indicates spore expressing H4K5,8,12R (green), or H3K9,14,18,23,27R (blue) as the sole source of H4 or H3 histones, respectively. The dashed circle indicates ctf4Δ spore expressing H4K5,8,12R (green), or H3K9,14,18,23,27R (blue) as the sole source of H4 or H3 histones, respectively.