Back to article: The role of hydrophobic matching on transmembrane helix packing in cells
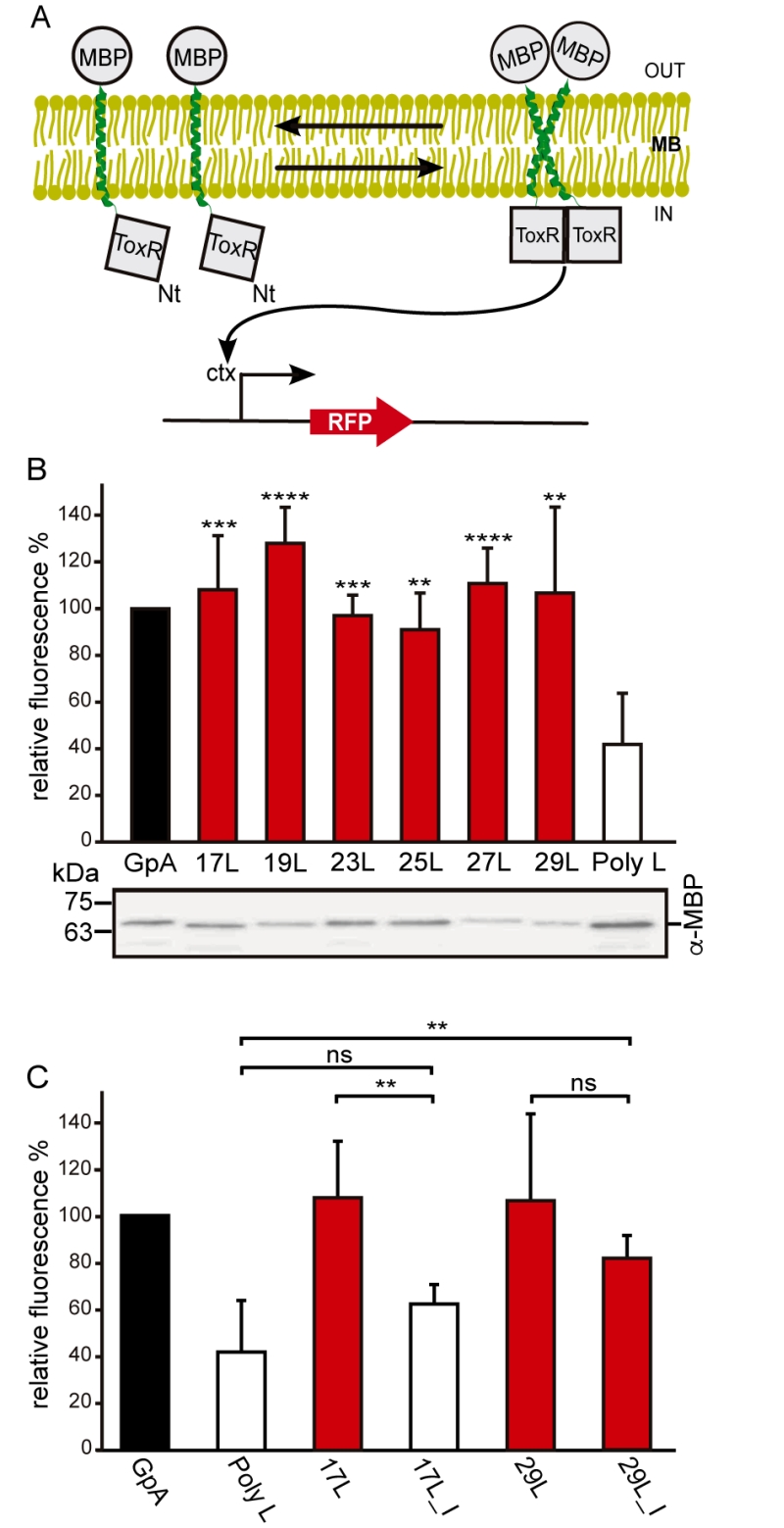
FIGURE 1: Homo-dimerization in E. coli membranes. (A) Schematic representation of ToxRED Assay. TM-driven oligomerization results in dimerization of ToxR transcriptional activator which, ultimately, drives the expression of the red fluorescence protein RFP (encoded under ctx promoter). The c-terminus maltose binding protein (MBP) located in the periplasm (OUT) allows growth of E. coli MM39 strain in M9 minimal media supplemented with 0.8% of maltose. (B) Mean relative fluorescence of chimera homo-oligomerization. Error bars denote standard deviation obtained from at least 6 independent experiments (p-values for the comparison with poly L: ** <0.01, *** <0.001, **** <0.0001). The color intensity-code was used to highlight dimerization (red) vs non-dimerization (white). The positive control (GpA homo-dimer) is shown in black. The α-MBP western blot under the bar graph shows chimera’s expression levels. (C) Contribution of Gly for the dimerization of TM chimeras. Mean relative fluorescence of 17L, 29L, 17L_I, and 29I_L chimeras homo-oligomerization. Error bars denote standard deviation obtained from at least 4 independent experiments (p-values for the comparison with poly L: ** <0.01, ns non-significant). The color intensity-code is used to highlight dimerization (red) vs non-dimerization (white). Positive control (GpA homo-dimer) is highlighted in black.