Back to article: A game of substrates: replication fork remodeling and its roles in genome stability and chemo-resistance
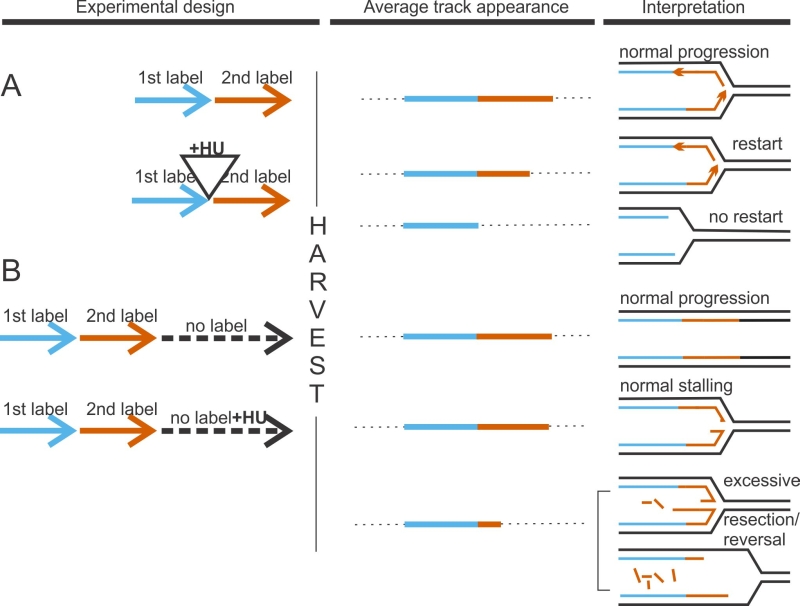
FIGURE 1: DNA fiber analysis detects faulty fork activity. In a “classic” experiment to measure fork restart (A), cells are sequentially pulse-labeled for 15-30 min each with two nucleoside analogs, typically, chloro-deoxyuridine and iodo-deoxyuridine. In test samples, incubation with a fork-arresting dose of hydroxyurea (HU), a ribonucleotide reductase inhibitor, is introduced between the labels. Failure to restart after HU manifests as preponderance of tracks containing only 1st label. One way to measure degradation of nascent strands at stalled forks (B), is to follow sequential pulse-labeling with two labels by a label-free chase (usually 4-6hrs). In test samples, HU is introduced during this time. Extensive loss of nascent DNA manifests as shortening of 2nd label segments in tracks labeled prior to HU addition.