Back to article: mIDH-associated DNA hypermethylation in acute myeloid leukemia reflects differentiation blockage rather than inhibition of TET-mediated demethylation
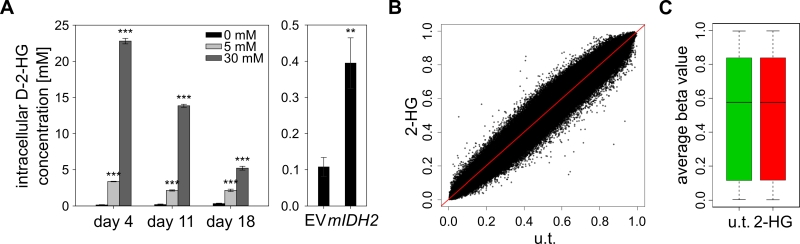
FIGURE 5: D-2-HG does not induce genomic hypermethylation in cultured cells. (A) Measurement of intracellular D-2-HG concentrations using an enzymatic conversion assay [50]. HL-60 cells were cultured in medium containing 5 or 30 mM synthetic D-2-HG and quantifications of intracellular concentrations were performed at the indicated time points. The right panel shows intracellular concentrations in HL-60 cells transduced with empty vector (EV) or mIDH2. Statistical significance was calculated using the two-sided Student’s t-test (*** P<0.001 and ** P<0.01). (B) Comparison of methylomes from D-2-HG treated (2-HG; 30 mM for 21 days) and untreated (u.t.) HL-60 cells by scatterplot. No significantly changed probes (adj. P<0.05) were found analyzing two biological replicates of each condition by EPIC methylation array. (C) Boxplot of average beta values of all probes in untreated and D-2-HG treated (30 mM for 21 days) HL-60 cells (P=0.17).
50. Balss J, Pusch S, Beck AC, Herold-Mende C, Krämer A, Thiede C, Buckel W, Langhans CD, Okun JG, Von Deimling A (2012). Enzymatic assay for quantitative analysis of (d)-2-hydroxyglutarate. Acta Neuropathol 124(6): 883-891. https://doi.org/10.1007/s00401-012-1060-y