Back to article: mIDH-associated DNA hypermethylation in acute myeloid leukemia reflects differentiation blockage rather than inhibition of TET-mediated demethylation
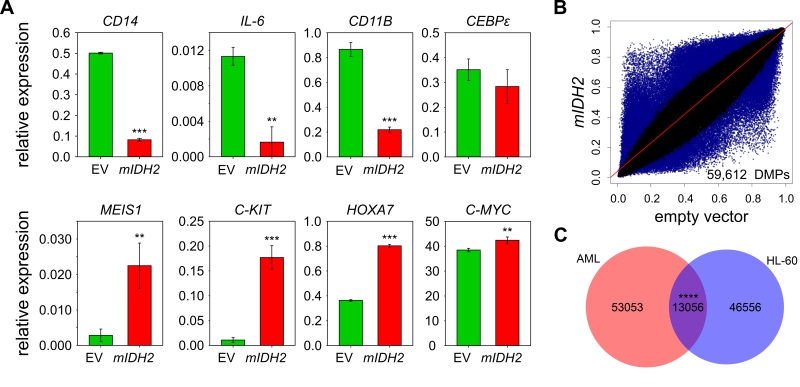
FIGURE 2: Genomic methylation profiles of mIDH AML patients and HL-60 cells expressing IDH2 R140Q are distinct. (A) Expression analysis of myeloid progenitor and differentiation genes by qRT-PCR in HL-60 cells transduced with empty vector (EV) or mIDH2. Expression is shown relative to ACTB transcript levels. Error bars indicate standard deviation (n=3). Statistical significance was calculated using a two-sided Student’s t-test (*** P<0.001, ** P<0.01). (B) Scatterplots comparing average beta values for all probes common to 450K and EPIC chip between HL-60 cells transduced with mIDH2 or empty vector. Each dot represents one probe with DMPs colored in blue. Two biological replicates were analyzed independently on the 450K chip per condition. (C) Venn diagram showing the overlap between DMPs found upon presence of mIDH in the AML patient dataset and the HL-60 cells. The overlap between the two groups was minor, but greater than expected by chance using the hypergeometric test (**** P<0.0001).